2D57
 
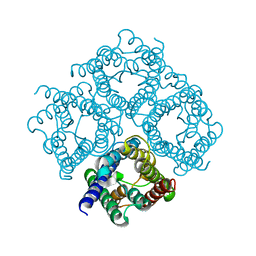 | | Double layered 2D crystal structure of AQUAPORIN-4 (AQP4M23) at 3.2 a resolution by electron crystallography | | Descriptor: | Aquaporin-4 | | Authors: | Hiroaki, Y, Tani, K, Kamegawa, A, Gyobu, N, Nishikawa, K, Suzuki, H, Walz, T, Sasaki, S, Mitsuoka, K, Kimura, K, Mizoguchi, A, Fujiyoshi, Y. | | Deposit date: | 2005-10-29 | | Release date: | 2006-01-31 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Implications of the Aquaporin-4 Structure on Array Formation and Cell Adhesion
J.Mol.Biol., 355, 2005
|
|
2DN2
 
 | | 1.25A resolution crystal structure of human hemoglobin in the deoxy form | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Park, S.-Y, Yokoyama, T, Shibayama, N, Shiro, Y, Tame, J.R. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 a resolution crystal structures of human haemoglobin in the oxy, deoxy and carbonmonoxy forms.
J.Mol.Biol., 360, 2006
|
|
2YN9
 
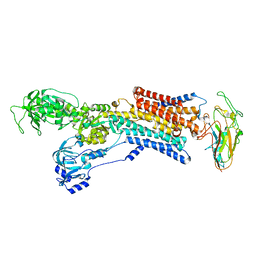 | | Cryo-EM structure of gastric H+,K+-ATPase with bound rubidium | | Descriptor: | POTASSIUM-TRANSPORTING ATPASE ALPHA CHAIN 1, POTASSIUM-TRANSPORTING ATPASE SUBUNIT BETA | | Authors: | Abe, K, Tani, K, Friedrich, T, Fujiyoshi, Y. | | Deposit date: | 2012-10-13 | | Release date: | 2012-11-07 | | Last modified: | 2014-07-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Cryo-Em Structure of Gastric H+,K+-ATPase with a Single Occupied Cation-Binding Site.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YQS
 
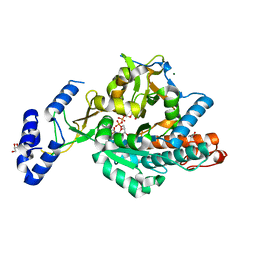 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the product-binding form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YVE
 
 | | Crystal structure of the methylene blue-bound form of the multi-drug binding transcriptional repressor CgmR | | Descriptor: | 3,7-BIS(DIMETHYLAMINO)PHENOTHIAZIN-5-IUM, CHLORIDE ION, GLYCEROL, ... | | Authors: | Itou, H, Shirakihara, Y, Tanaka, I. | | Deposit date: | 2007-04-12 | | Release date: | 2008-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Multidrug Binding Repressor Corynebacteriumglutamicum CgmR in Complex with Inducers and with an Operator
J.Mol.Biol., 403, 2010
|
|
2YQJ
 
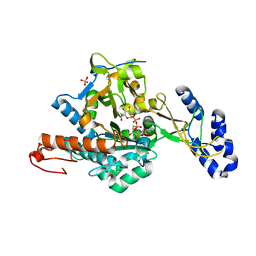 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the reaction-completed form | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YQC
 
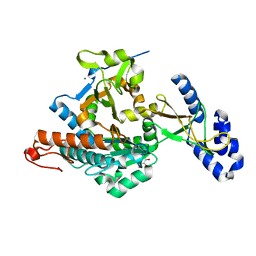 | | Crystal Structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the apo-like form | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-N-acetylglucosamine pyrophosphorylase | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2YVH
 
 | |
2Z1O
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-05-10 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZMU
 
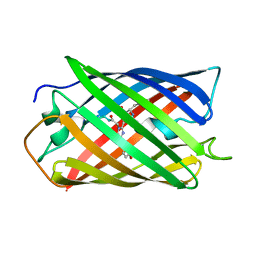 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 9.1 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
2Z6X
 
 | | Crystal structure of 22G, the wild-type protein of the photoswitchable GFP-like protein Dronpa | | Descriptor: | photochromic protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AG0
 
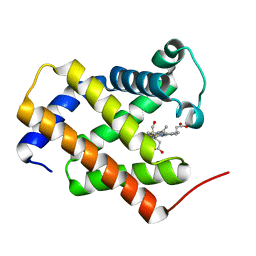 | | Crystal structure of carbonmonoxy human cytoglobin | | Descriptor: | CARBON MONOXIDE, Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sawai, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2010-03-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the carbon monoxide complex of human cytoglobin
Proteins, 79, 2011
|
|
2ZZA
 
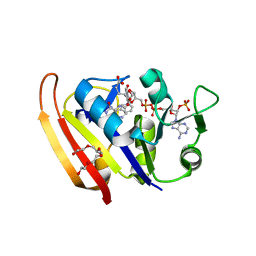 | | Moritella profunda Dihydrofolate reductase complex with NADP+ and Folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hata, K, Tanaka, T, Murakami, C, Ohmae, E, Gekko, K, Shiro, Y, Akasaka, K. | | Deposit date: | 2009-02-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Moritella profunda Dihydrofolate reductase complex with NADP+ and Folate
To be Published
|
|
3A5P
 
 | | Crystal structure of hemagglutinin | | Descriptor: | Haemagglutinin I | | Authors: | Watanabe, N, Sakai, N, Nakamura, T, Nabeshima, Y, Kouno, T, Mizuguchi, M, Kawano, K. | | Deposit date: | 2009-08-10 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Structure of Physarum polycephalum hemagglutinin I suggests a minimal carbohydrate recognition domain of legume lectin fold
J.Mol.Biol., 405, 2011
|
|
2ZF3
 
 | | Crystal Structure of VioE | | Descriptor: | Hypothetical protein VioE | | Authors: | Hirano, S, Shiro, Y, Nagano, S. | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of VioE, a key player in the construction of the molecular skeleton of violacein
To be Published
|
|
2ZF4
 
 | |
2Z6Y
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Z33
 
 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
2Z6Z
 
 | | Crystal structure of a photoswitchable GFP-like protein Dronpa in the bright-state | | Descriptor: | Fluorescent protein Dronpa | | Authors: | Kikuchi, A, Jeyakanthan, J, Taka, J, Shiro, Y, Mizuno, H, Miyawaki, A. | | Deposit date: | 2007-08-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-dependent regulation of structural flexibility in a photochromic fluorescent protein.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZMW
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 6.0 | | Descriptor: | Fluorescent protein | | Authors: | Kikuchi, A, Fukumura, E, Karasawa, S, Mizuno, H, Miyawaki, A, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-04-21 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of a Thiazoline-Containing Chromophore in an Orange Fluorescent Protein, Monomeric Kusabira Orange
Biochemistry, 47, 2008
|
|
2E89
 
 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
2DC3
 
 | | Crystal structure of human cytoglobin at 1.68 angstroms resolution | | Descriptor: | ACETIC ACID, Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Sawai, H, Kawada, N, Yoshizato, K, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | High-resolution structure of human cytoglobin: identification of extra N- and C-termini and a new dimerization mode.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2CFM
 
 | |
2D2A
 
 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
2AT9
 
 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | 3-[[3-METHYLPHOSPHONO-GLYCEROLYL]PHOSPHONYL]-[1,2-DI[2,6,10,14-TETRAMETHYL-HEXADECAN-16-YL]GLYCEROL, BACTERIORHODOPSIN, RETINAL | | Authors: | Mitsuoka, K, Hirai, T, Murata, K, Miyazawa, A, Kidera, A, Kimura, Y, Fujiyoshi, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | The structure of bacteriorhodopsin at 3.0 A resolution based on electron crystallography: implication of the charge distribution.
J.Mol.Biol., 286, 1999
|
|
