1O9O
 
 | |
1O9P
 
 | |
1OCL
 
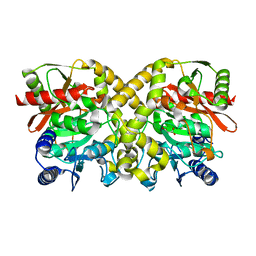 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, MALONIC ACID | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
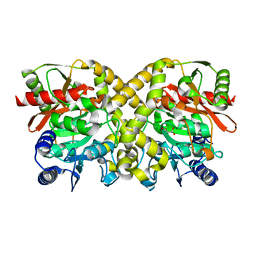 | |
1Q1O
 
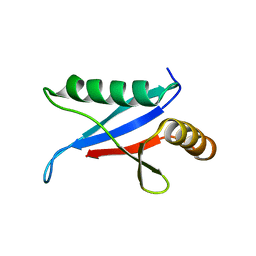 | | Solution Structure of the PB1 Domain of Cdc24p (Long Form) | | Descriptor: | Cell division control protein 24 | | Authors: | Yoshinaga, S, Kohjima, M, Ogura, K, Yokochi, M, Takeya, R, Ito, T, Sumimoto, H, Inagaki, F. | | Deposit date: | 2003-07-22 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The PB1 domain and the PC motif-containing region are structurally similar protein binding modules
EMBO J., 22, 2003
|
|
4ZZ8
 
 | | X-ray crystal structure of chitosan-binding module 2 in complex with chitotriose derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Glucanase/chitosanase, ... | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
3SQG
 
 | | Crystal structure of a methyl-coenzyme M reductase purified from Black Sea mats | | Descriptor: | (17[2]S)-17[2]-methylthio-coenzyme F43, 1-THIOETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shima, S, Krueger, M, Weinert, T, Demmer, U, Thauer, R.K, Ermler, U. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically.
Nature, 481, 2011
|
|
1O9Q
 
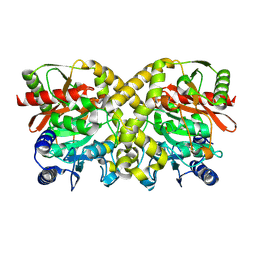 | |
1OCK
 
 | |
1OCH
 
 | |
1OBJ
 
 | |
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | Descriptor: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | Authors: | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | Deposit date: | 2003-02-08 | | Release date: | 2003-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | Descriptor: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
1O9N
 
 | |
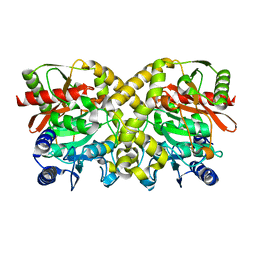
1OBI
 
 | |
4ZY9
 
 | | X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 | | Descriptor: | Glucanase/chitosanase | | Authors: | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | Deposit date: | 2015-05-21 | | Release date: | 2016-04-13 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
1QLG
 
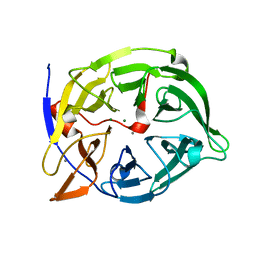 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | Descriptor: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | Authors: | Shin, S, Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-08-31 | | Release date: | 2000-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
3A31
 
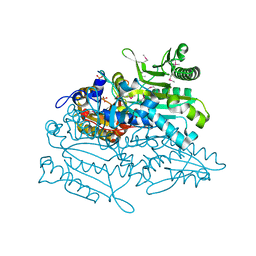 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix (selenomethionine derivative) | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
3A32
 
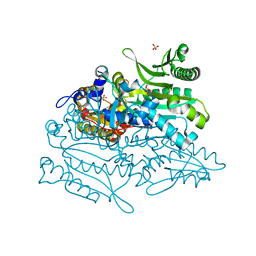 | | Crystal structure of putative threonyl-tRNA synthetase ThrRS-1 from Aeropyrum pernix | | Descriptor: | Probable threonyl-tRNA synthetase 1, SULFATE ION, ZINC ION | | Authors: | Shimizu, S, Juan, E.C.M, Miyashita, Y, Sato, Y, Hoque, M.M, Suzuki, K, Yogiashi, M, Tsunoda, M, Dock-Bregeon, A.-C, Moras, D, Sekiguchi, T, Takenaka, A. | | Deposit date: | 2009-06-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two complementary enzymes for threonylation of tRNA in crenarchaeota: crystal structure of Aeropyrum pernix threonyl-tRNA synthetase lacking a cis-editing domain
J.Mol.Biol., 394, 2009
|
|
6K3I
 
 | |
2KTD
 
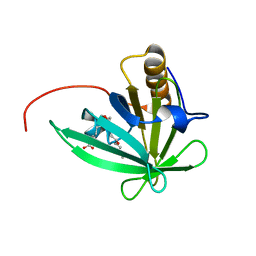 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
3VJN
 
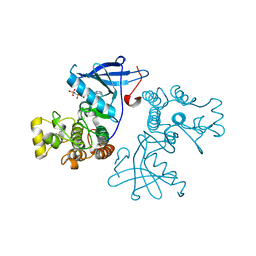 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
1EZW
 
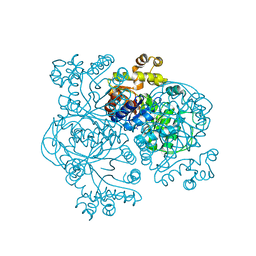 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F07
 
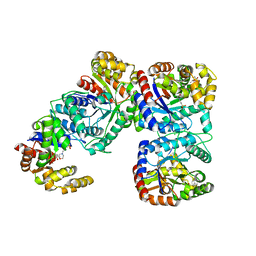 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
