2CUB
 
 | | Solution structure of the SH3 domain of the human cytoplasmic protein Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Ohnishi, S, Kigawa, T, Sato, M, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the human cytoplasmic protein Nck1
To be Published
|
|
2QZK
 
 | | Crystal structure of human Beta Secretase complexed with I21 | | Descriptor: | 2-[(5R)-5-amino-5-methyl-4,16-dioxo-14-phenyl-3-oxa-15-azatricyclo[15.3.1.1~7,11~]docosa-1(21),7(22),8,10,12,14,17,19-octaen-19-yl]benzonitrile, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategies toward improving the brain penetration of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2DMV
 
 | | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch) | | Descriptor: | Itchy homolog E3 ubiquitin protein ligase | | Authors: | Ohnishi, S, Paakkonen, K, Tochio, N, Tomizawa, T, Koshiba, S, Inoue, M, Guntert, P, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch)
To be Published
|
|
2DMW
 
 | | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1 | | Descriptor: | synaptobrevin-like 1 variant | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1
To be Published
|
|
2DMX
 
 | | Solution structure of the J domain of DnaJ homolog subfamily B member 8 | | Descriptor: | DnaJ homolog subfamily B member 8 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the J domain of DnaJ homolog subfamily B member 8
To be Published
|
|
1V8L
 
 | | Structure Analysis of the ADP-ribose pyrophosphatase complexed with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-10 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8R
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8Y
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E86Q mutant, complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8T
 
 | | Crystal Structure analysis of the ADP-ribose pyrophosphatase complexed with ribose-5'-phosphate and Zn | | Descriptor: | ADP-ribose pyrophosphatase, RIBOSE-5-PHOSPHATE, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8I
 
 | | Crystal Structure Analysis of the ADP-ribose pyrophosphatase | | Descriptor: | ADP-ribose pyrophosphatase | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-09 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8W
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant, complexed with SO4 and Zn | | Descriptor: | ADP-ribose pyrophosphatase, SULFATE ION, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
2ONW
 
 | |
1V8N
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase complexed with Zn | | Descriptor: | ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8V
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E86Q mutant, complexed with ADP-ribose and Mg | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8U
 
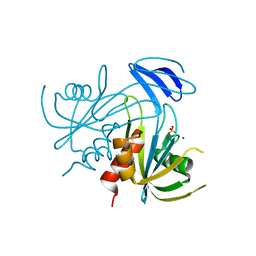 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant with SO4 and Mg | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, SULFATE ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8M
 
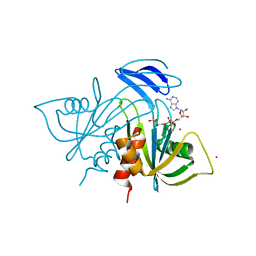 | | Crystal structure analysis of ADP-ribose pyrophosphatase complexed with ADP-ribose and Gd | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, GADOLINIUM ATOM | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
2ON9
 
 | |
2QZL
 
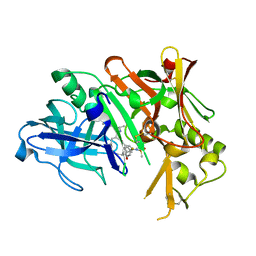 | | Crystal Structure of human Beta Secretase complexed with IXS | | Descriptor: | Beta-secretase 1, N-[(1S)-1-benzyl-2-{[(1S)-2-(isobutylamino)-1-methyl-2-oxoethyl]amino}ethyl]-N'-[(1R)-1-(4-fluorophenyl)ethyl]-5-[methyl(methylsulfonyl)amino]isophthalamide | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BACE-1 inhibition by a series of psi[CH2NH] reduced amide isosteres
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2ONA
 
 | |
2ONV
 
 | |
2KWY
 
 | | Structure of G61-101 | | Descriptor: | V-type proton ATPase subunit G | | Authors: | Rishikesan, S, Gruber, G. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of G61-101
To be Published
|
|
3B1B
 
 | |
3VOC
 
 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
2OAH
 
 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-[(1S,2S)-2-AMINO-1-(3-THIENYLMETHYL)HEXYL]-2-({[(1S,2S)-2-METHYLCYCLOPROPYL]METHYL}AMINO)-6-[METHYL(METHYLSULFONYL)AMINO]ISONICOTINAMIDE | | Authors: | Munshi, S. | | Deposit date: | 2006-12-15 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and SAR of isonicotinamide BACE-1 inhibitors that bind beta-secretase in a N-terminal 10s-loop down conformation.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
