7RXS
 
 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXT
 
 | | Crystal of BRD4(D1) with 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 2-[(3R)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RXR
 
 | | Crystal Structure of BRD4(D1) with 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-(4-bromophenyl)-1-(piperidin-4-yl)-1H-imidazol-5-yl]-N-(3,5-dimethylphenyl)pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
7RWN
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWP
 
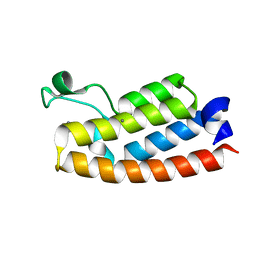 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
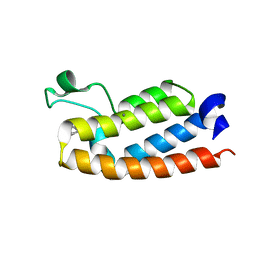 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
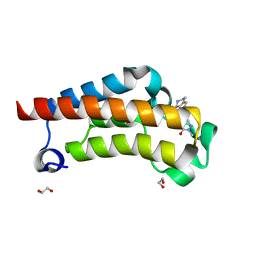 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
6VW1
 
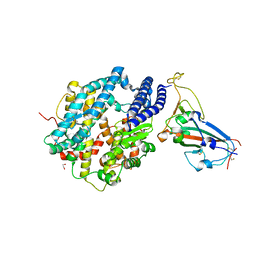 | | Structure of SARS-CoV-2 chimeric receptor-binding domain complexed with its receptor human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Ye, G, Shi, K, Wan, Y.S, Aihara, H, Li, F. | | Deposit date: | 2020-02-18 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural basis of receptor recognition by SARS-CoV-2.
Nature, 581, 2020
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
3LMX
 
 | | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Purpero, V.M, Lipscomb, J.D, Shi, K. | | Deposit date: | 2010-02-01 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tyrosine 447 of Protocatechuate 34,-Dioxygenase Controls Efficient Progress Through Catalysis
To be Published
|
|
6VK7
 
 | | Crystal Structure of reduced Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | Descriptor: | FE (III) ION, Methane monooxygenase, Methane monooxygenase component A alpha chain | | Authors: | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2020-01-18 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
7S6R
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with H5A mutation | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6S
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL1 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6Q
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S7H
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit DBL2 | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Methane monooxygenase beta chain, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
7S6T
 
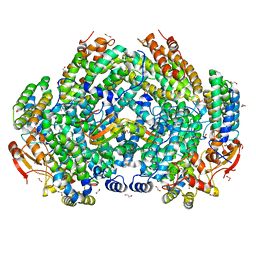 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit H33A | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, FE (III) ION, ... | | Authors: | Johns, J.C, Banerjee, R, Semonis, M.M, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structures of Methane Monooxygenase Hydroxylase Complexes with Variants of Its Regulatory Component: Correlations with Altered Reaction Cycle Dynamics.
Biochemistry, 61, 2022
|
|
5EJK
 
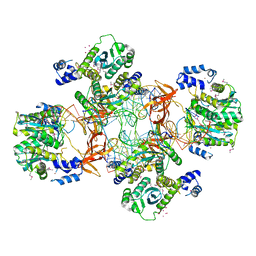 | | Crystal structure of the Rous sarcoma virus intasome | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*T)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*CP*TP*C)-3'), ... | | Authors: | Yin, Z, Shi, K, Banerjee, S, Aihara, H. | | Deposit date: | 2015-11-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of the Rous sarcoma virus intasome.
Nature, 530, 2016
|
|
7U0N
 
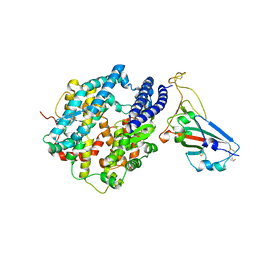 | | Crystal structure of chimeric omicron RBD (strain BA.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Geng, Q, Shi, K, Ye, G, Zhang, W, Aihara, H, Li, F. | | Deposit date: | 2022-02-18 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Basis for Human Receptor Recognition by SARS-CoV-2 Omicron Variant BA.1.
J.Virol., 96, 2022
|
|
7TGR
 
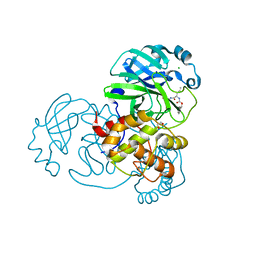 | | Structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Esler, M.A, Shi, K, Aihara, H, Harris, R.S. | | Deposit date: | 2022-01-09 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Gain-of-Signal Assays for Probing Inhibition of SARS-CoV-2 M pro /3CL pro in Living Cells.
Mbio, 13, 2022
|
|
3I41
 
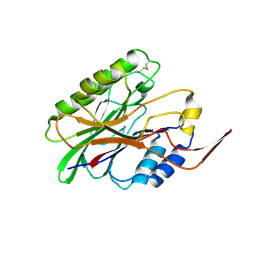 | |
3I48
 
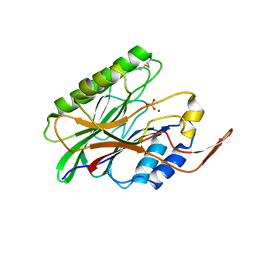 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound magnesium ions | | Descriptor: | Beta-hemolysin, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
to be published, 2009
|
|
3I5V
 
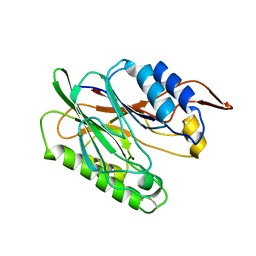 | |
419D
 
 | |
3I46
 
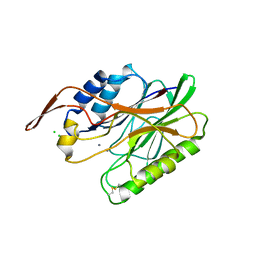 | | Crystal structure of beta toxin from Staphylococcus aureus F277A, P278A mutant with bound calcium ions | | Descriptor: | Beta-hemolysin, CALCIUM ION, CHLORIDE ION | | Authors: | Huseby, M, Shi, K, Kruse, A.C, Ohlendorf, D.H. | | Deposit date: | 2009-07-01 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and biological functions of beta toxin from Staphylococcus aureus: Role of the hydrophobic beta hairpin in virulence
To be Published
|
|
