8OZV
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex with 2,2-difluoroacetophenone | | Descriptor: | 2,2-bis(fluoranyl)-1-phenyl-ethanone, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8OZW
 
 | | Imine Reductase from Ajellomyces dermatitidis in complex NADPH4 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2023-05-09 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the imine reductase from Ajellomyces dermatitidis in three crystal forms.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6EOD
 
 | |
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q58
 
 | |
8QC3
 
 | |
6TOE
 
 | |
3IY9
 
 | | Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L16, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6SLE
 
 | | Structure of Reductive Aminase from Neosartorya fumigata in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
7X2Y
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase in complex with NAD+ and 3-Hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, 4,5-dihydroxyphthalate dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
6SMY
 
 | |
6SMZ
 
 | |
8QC6
 
 | |
1DQ7
 
 | | THREE-DIMENSIONAL STRUCTURE OF A NEUROTOXIN FROM RED SCORPION (BUTHUS TAMULUS) AT 2.2A RESOLUTION. | | Descriptor: | NEUROTOXIN | | Authors: | Sharma, M, Yadav, S, Karthikeyan, S, Kumar, S, Paramasivam, M, Srinivasan, A, Singh, T.P. | | Deposit date: | 1999-12-30 | | Release date: | 2000-12-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional Structure of a Neurotoxin from Red Scorpion (Buthus tamulus) at 2.2A Resolution
To be Published
|
|
6SKX
 
 | | Structure of Reductive Aminase from Neosartorya fumigata | | Descriptor: | Oxidoreductase, putative | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2019-08-16 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Asymmetric synthesis of primary amines catalyzed by thermotolerant fungal reductive aminases.
Chem Sci, 11, 2020
|
|
6SM7
 
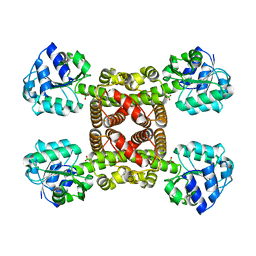 | | Crystal structure of SLA Reductase YihU from E. Coli | | Descriptor: | 3-sulfolactaldehyde reductase, BORIC ACID | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dynamic Structural Changes Accompany the Production of Dihydroxypropanesulfonate by Sulfolactaldehyde Reductase
Acs Catalysis, 2020
|
|
6NU3
 
 | | Structural insights into unique features of the human mitochondrial ribosome recycling | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into unique features of the human mitochondrial ribosome recycling.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6TO4
 
 | |
6NU2
 
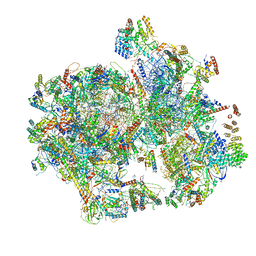 | | Structural insights into unique features of the human mitochondrial ribosome recycling | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2019-01-30 | | Release date: | 2019-04-17 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into unique features of the human mitochondrial ribosome recycling.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6EOH
 
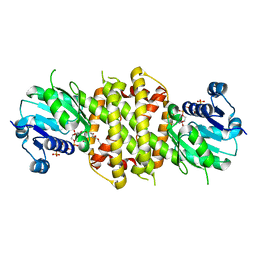 | | Reductive Aminase from Aspergillus terreus in complex with NADPH and ethyl levulinate | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductive Aminase, ethyl levulinate | | Authors: | Sharma, M, Grogan, G, Mangas-Sanchez, J, Turner, N.J. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mechanism for Reductive Amination Catalyzed by Fungal Reductive Aminases
Acs Catalysis, 2018
|
|
6EOI
 
 | | Reductive Aminase from Aspergillus terreus in complex with NADPH and ethyl-5-oxohexanoate | | Descriptor: | MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Reductive Aminase, ... | | Authors: | Sharma, M, Mangas-Sanchez, J, Turner, N.J, Grogan, G. | | Deposit date: | 2017-10-09 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Mechanism for Reductive Amination Catalyzed by Fungal Reductive Aminases
Acs Catalysis, 2018
|
|
7WZD
 
 | | Crystal Structure of cis-4,5-dihydrodiol phthalate dehydrogenase from Comamonas testosteroni KF1 | | Descriptor: | 4,5-dihydroxyphthalate dehydrogenase, GLYCEROL | | Authors: | Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational flexibility enables catalysis of phthalate cis-4,5-dihydrodiol dehydrogenase.
Arch.Biochem.Biophys., 727, 2022
|
|
1X18
 
 | | Contact sites of ERA GTPase on the THERMUS THERMOPHILUS 30S SUBUNIT | | Descriptor: | 30S ribosomal protein S11, 30S ribosomal protein S18, 30S ribosomal protein S2, ... | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
1X1L
 
 | | Interaction of ERA,a GTPase protein, with the 3'minor domain of the 16S rRNA within the THERMUS THERMOPHILUS 30S subunit. | | Descriptor: | GTP-binding protein era, RNA (130-MER) | | Authors: | Sharma, M.R, Barat, C, Agrawal, R.K. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Interaction of Era with the 30S Ribosomal Subunit Implications for 30S Subunit Assembly
Mol.Cell, 18, 2005
|
|
6VLZ
 
 | | Structure of the human mitochondrial ribosome-EF-G1 complex (ClassI) | | Descriptor: | 12s rRNA, 16s rRNA, 28S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Koripella, R.K, Agrawal, R.K. | | Deposit date: | 2020-01-27 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of the human mitochondrial ribosome bound to EF-G1 reveal distinct features of mitochondrial translation elongation.
Nat Commun, 11, 2020
|
|
