1V6D
 
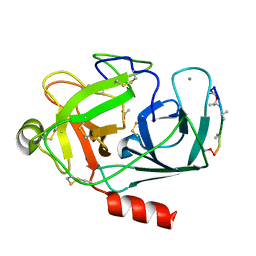 | |
1DII
 
 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
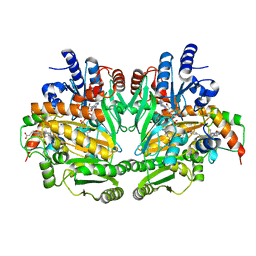
1DIQ
 
 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
1UHB
 
 | | Crystal structure of porcine alpha trypsin bound with auto catalyticaly produced native peptide at 2.15 A resolution | | Descriptor: | 9-mer peptide from Trypsin, ACETATE ION, CALCIUM ION, ... | | Authors: | Pattabhi, V, Syed Ibrahim, B, Shamaladevi, N. | | Deposit date: | 2003-06-27 | | Release date: | 2004-07-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Trypsin activity reduced by an autocatalytically produced nonapeptide.
J.Biomol.Struct.Dyn., 21, 2004
|
|
