6BRT
 
 | | F-box protein CTH with hydrolase | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, D3-CTH-D14-D-ring | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-12-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6BRQ
 
 | | Crystal structure of rice ASK1-D3 ubiquitin ligase complex crystal form 3 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6DSZ
 
 | |
6BRP
 
 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6BRO
 
 | |
7UPV
 
 | |
7SA1
 
 | | LRR-F-Box plant ubiquitin ligase | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, F-box/LRR-repeat MAX2 homolog, ... | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2021-09-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | A conformational switch in the SCF-D3/MAX2 ubiquitin ligase facilitates strigolactone signalling.
Nat.Plants, 8, 2022
|
|
6X24
 
 | | Structural basis of plant blue light photoreceptor | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Palayam, M, Ganapathy, J, Guercio, M.A, Shabek, N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into photoactivation of plant Cryptochrome-2.
Commun Biol, 4, 2021
|
|
7K2Z
 
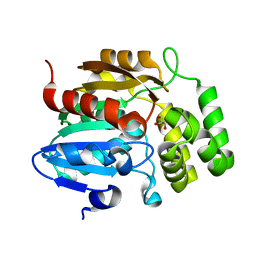 | | Crystal structure of Pisum sativum KAI2 Apo form | | Descriptor: | GLYCEROL, PsKAI2B protein | | Authors: | Guercio, A.M, Shabek, N. | | Deposit date: | 2020-09-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural and functional analyses explain Pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception.
Commun Biol, 5, 2022
|
|
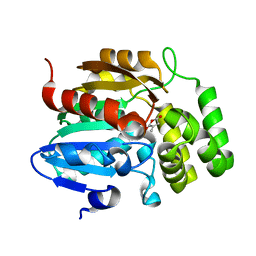
7K38
 
 | |
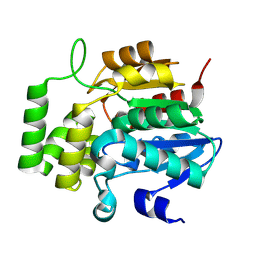
8VCZ
 
 | |
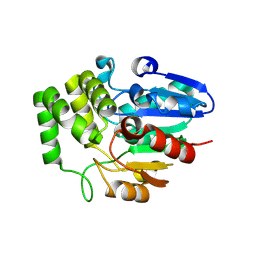
8VD3
 
 | |
8VCD
 
 | |
8VCA
 
 | | Crystal Structure of plant Carboxylesterase 15 | | Descriptor: | Carboxylesterase 15, GLYCEROL | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
8VD1
 
 | | Crystal Structure of KAI2 from Oryza sativa with MPD | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable esterase D14L | | Authors: | Gilio, A.K, Shabek, N, Guercio, A.M, Pawlak, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural insights into rice KAI2 receptor provide functional implications for perception and signal transduction.
J.Biol.Chem., 300, 2024
|
|
8VCE
 
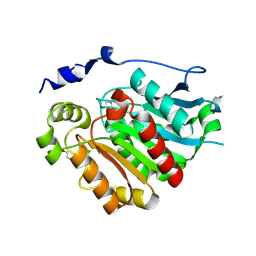 | | Crystal Structure of plant Carboxylesterase 20 | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Probable carboxylesterase 120 | | Authors: | Palayam, M, Shabek, N. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into strigolactone catabolism by carboxylesterases reveal a conserved conformational regulation.
Nat Commun, 15, 2024
|
|
