2BAW
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of the PPAR-beta/delta Ligand Binding Domain Model Reveals Why It Exhibits the Activated Form
Mol.Cell, 21, 2006
|
|
2GR1
 
 | |
2B7B
 
 | | Yeast guanine nucleotide exchange factor eEF1Balpha K205A mutant in complex with eEF1A and GDP | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE, elongation factor-1 beta | | Authors: | Pittman, Y.R, Valente, L, Jeppesen, M.G, Andersen, G.R, Patel, S, Kinzy, T.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
J.Biol.Chem., 281, 2006
|
|
2B7C
 
 | | Yeast guanine nucleotide exchange factor eEF1Balpha K205A mutant in complex with eEF1A | | Descriptor: | Elongation factor 1-alpha, elongation factor-1 beta | | Authors: | Pittman, Y.R, Valente, L, Jeppesen, M.G, Andersen, G.R, Patel, S, Kinzy, T.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
J.Biol.Chem., 281, 2006
|
|
2GR3
 
 | |
4E7T
 
 | | The structure of T6 bovine insulin | | Descriptor: | Insulin A chain, Insulin B chain, ZINC ION | | Authors: | Harris, P, Frankaer, C.G, Knudsen, M.V. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4AF2
 
 | | C61S mutant of thiol peroxidase form E. coli. | | Descriptor: | THIOL PEROXIDASE | | Authors: | Beckham, K.S.H, Roe, A.J, Byron, O, Gabrielsen, M. | | Deposit date: | 2012-01-16 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structure of an Orthorhombic Crystal Form of a `Forced Reduced' Thiol Peroxidase Reveals Lattice Formation Aided by the Presence of the Affinity Tag
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2GR2
 
 | |
4E7U
 
 | | The structure of T3R3 bovine insulin | | Descriptor: | Insulin A chain, Insulin B chain, THIOCYANATE ION, ... | | Authors: | Harris, P, Frankaer, C.G, Knudsen, M.V. | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structures of T(6), T(3)R(3) and R(6) bovine insulin: combining X-ray diffraction and absorption spectroscopy.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1SQM
 
 | | STRUCTURE OF [R563A] LEUKOTRIENE A4 HYDROLASE | | Descriptor: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | Authors: | Tholander, F.O.T, Rudberg, P.C, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | Deposit date: | 2004-03-19 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leukotriene A4 hydrolase: identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates
J.Biol.Chem., 279, 2004
|
|
1WLE
 
 | | Crystal Structure of mammalian mitochondrial seryl-tRNA synthetase complexed with seryl-adenylate | | Descriptor: | SERYL ADENYLATE, Seryl-tRNA synthetase | | Authors: | Chimnaronk, S, Jeppesen, M.G, Suzuki, T, Nyborg, J, Watanabe, K. | | Deposit date: | 2004-06-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dual-mode recognition of noncanonical tRNAs(Ser) by seryl-tRNA synthetase in mammalian mitochondria
Embo J., 24, 2005
|
|
1B5K
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1JUE
 
 | | 1.8 A resolution structure of native lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1GDU
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, SULFATE ION, TRYPSIN | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-29 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GDN
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-LYS, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1B6X
 
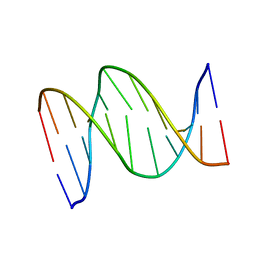 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE GUANINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 4 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3' | | Authors: | Cullinan, D, Johnson, F, Grollman, A.P, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyguanosine.
Biochemistry, 36, 1997
|
|
1JQV
 
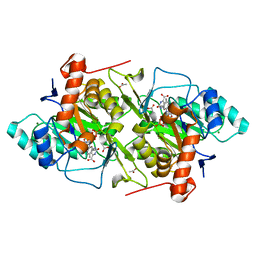 | | The K213E mutant of Lactococcus lactis Dihydroorotate dehydrogenase A | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase A, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1GDQ
 
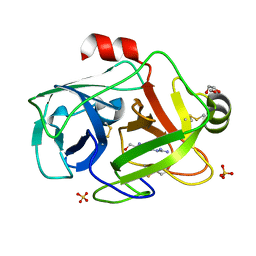 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2AWH
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-01 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2B50
 
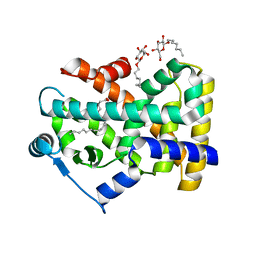 | | Human Nuclear Receptor-Ligand Complex 2 | | Descriptor: | CALCIUM ION, Peroxisome proliferator activated receptor delta, VACCENIC ACID, ... | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-09-27 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant Human PPAR-beta/delta Ligand-binding Domain is Locked in an Activated Conformation by Endogenous Fatty Acids
J.Mol.Biol., 356, 2006
|
|
2CHK
 
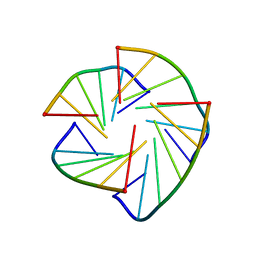 | | NMR structure of TLLLLT quadruplex | | Descriptor: | 5'-D(*T LCG LCG LCG LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
2CHJ
 
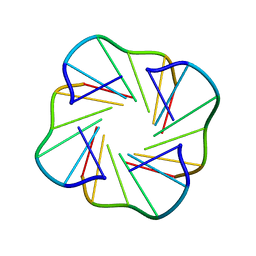 | | NMR structure of TGLGLT quadruplex | | Descriptor: | 5'-D(*TP*G LCGP*G LCGP*TP)-3' | | Authors: | Nielsen, J.T, Petersen, M. | | Deposit date: | 2006-03-15 | | Release date: | 2006-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structures of Lna (Locked Nucleic Acid) Modified Quadruplexes
Nucleic Acids Res., 34, 2006
|
|
1JUB
 
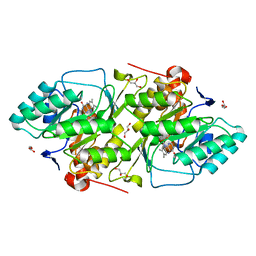 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1JQX
 
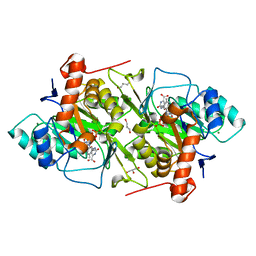 | | The R57A mutant of Lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-09 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
1FY4
 
 | | FUSARIUM OXYSPORUM TRYPSIN AT ATOMIC RESOLUTION | | Descriptor: | GLY-ALA-ARG, GLYCEROL, SULFATE ION, ... | | Authors: | Rypniewski, W.R, Oestergaard, P, Noerregaard-Madsen, M, Dauter, M, Wilson, K.S. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.81 Å) | | Cite: | Fusarium oxysporum trypsin at atomic resolution at 100 and 283 K: a study of ligand binding.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
