4BAT
 
 | |
4AN4
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, GLYCOSYL TRANSFERASE | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-15 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4BO0
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(4-methoxy-1- methylindazol-3-yl)-3-(2-methoxyphenyl)urea at 2.4A resolution | | Descriptor: | 1-(4-methoxy-1-methyl-indazol-3-yl)-3-(2-methoxyphenyl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO7
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2,3-dihydro-1H-inden- 5-yl)tetrazolo(1,5-b)pyridazin-6-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2,3-dihydro-1H-inden-5-yl)tetrazolo[1,5-b]pyridazin-6-amine | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO4
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-methoxyphenyl)-3,4- dihydro-2H-quinoline-1-carboxamide at 2.7A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-methoxyphenyl)-3,4-dihydro-2H-quinoline-1-carboxamide | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNX
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 6-(4-(2-chloroanilino)- 1H-quinazolin-2-ylidene)cyclohexa-2, 4-dien-1-one at 2.3A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 6-[4-(2-chloroanilino)-1H-quinazolin-2-ylidene]cyclohexa-2,4-dien-1-one | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO3
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-(3-(trifluoromethyl) anilino)pyridine-3-sulfonamide at 2.5A resolution | | Descriptor: | 2-(3-(trifluoromethyl)anilino)pyridine-3-sulfonamide, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNU
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 2-phenyl-4-(1,2,4- triazol-4-yl)quinazoline at 2.0A resolution | | Descriptor: | 2-phenyl-4-(1,2,4-triazol-4-yl)quinazoline, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO1
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(4-chloro-2,5- dimethoxyphenyl)quinoline-8-carboxamide at 2.2A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(4-CHLORO-2,5-DIMETHOXYPHENYL)QUINOLINE-8-CARBOXAMIDE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNW
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with an unknown ligand at 1. 6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BNV
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(2-chlorophenyl)-3-(1- methylbenzimidazol-2-yl)urea at 2.5A resolution | | Descriptor: | 1-(2-chlorophenyl)-3-(1-methylbenzimidazol-2-yl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in Resolution
Acs Chem.Biol., 8, 2013
|
|
4BAZ
 
 | |
4A7K
 
 | | Bifunctional Aldos-2-ulose dehydratase | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-14 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4A7Z
 
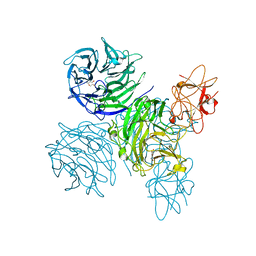 | | Complex of bifunctional aldos-2-ulose dehydratase with the reaction intermediate ascopyrone M | | Descriptor: | ALDOS-2-ULOSE DEHYDRATASE, Ascopyrone M, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4BO2
 
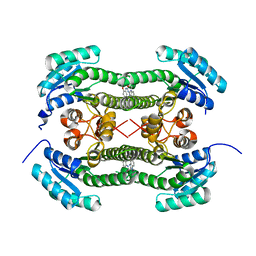 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 1-(1-ethylbenzimidazol-2- yl)-3-(2-methoxyphenyl)urea at 1.9A resolution | | Descriptor: | 1-(1-ethylbenzimidazol-2-yl)-3-(2-methoxyphenyl)urea, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4BO5
 
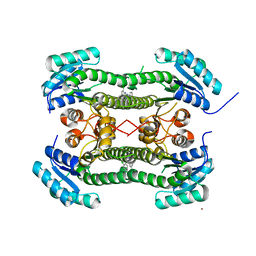 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2-chlorophenyl)-4- pyrrol-1-yl-1,3,5-triazin-2-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2-chlorophenyl)-4-pyrrol-1-yl-1,3,5-triazin-2-amine, NICKEL (II) ION | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4C5Z
 
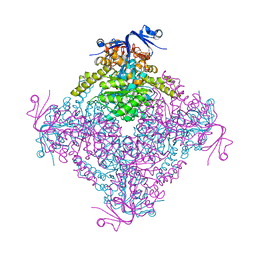 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4A7Y
 
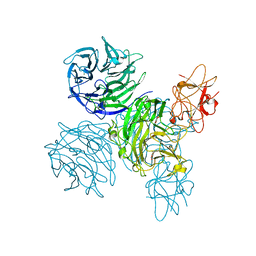 | | Active site metal depleted aldos-2-ulose dehydratase | | Descriptor: | 1,5-anhydro-D-fructose, ALDOS-2-ULOSE DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Claesson, M, Lindqvist, Y, Madrid, S, Sandalova, T, Fiskesund, R, Yu, S, Schneider, G. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bifunctional Aldos-2-Ulose Dehydratase/Isomerase from Phanerochaete Chrysosporium with the Reaction Intermediate Ascopyrone M.
J.Mol.Biol., 417, 2012
|
|
4BO9
 
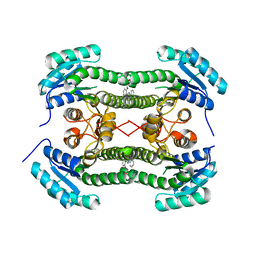 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with 5-(2-(furan-2-ylmethoxy) phenyl)-2-phenyltetrazole at 2.9A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, 5-[2-(FURAN-2-YLMETHOXY)PHENYL]-2-PHENYLTETRAZOLE | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4AMB
 
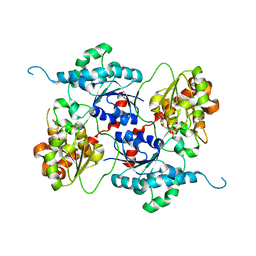 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4AMG
 
 | | Crystal structure of the glycosyltransferase SnogD from Streptomyces nogalater | | Descriptor: | SNOGD | | Authors: | Claesson, M, Siitonen, V, Dobritzsch, D, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2012-03-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of the Glycosyltransferase Snogd from the Biosynthetic Pathway of the Nogalamycin in Streptomyces Nogalater.
FEBS J., 279, 2012
|
|
4AIV
 
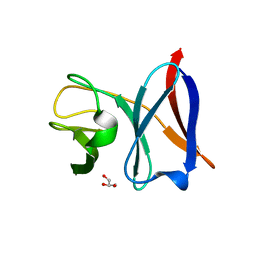 | | Crystal Structure of putative NADH-dependent nitrite reductase small subunit from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PROBABLE NITRITE REDUCTASE [NAD(P)H] SMALL SUBUNIT NIRD | | Authors: | Izumi, A, Schnell, R, Schneider, G. | | Deposit date: | 2012-02-15 | | Release date: | 2012-09-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Nird, the Small Subunit of the Nitrite Reductase Nirbd from Mycobacterium Tuberculosis, at 2.0 Angstrom Resolution
Proteins, 80, 2012
|
|
6TJA
 
 | | Crystal structure of the SVS_A2 protein (W79F,G83L mutant) from ancestral sequence reconstruction at 2.27 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, SVS_variant_AS1 | | Authors: | Rudraraju, R, Schnell, R, Schneider, G. | | Deposit date: | 2019-11-25 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Engineering of Ancestors as a Tool to Elucidate Structure, Mechanism, and Specificity of Extant Terpene Cyclase.
J.Am.Chem.Soc., 143, 2021
|
|
6TIV
 
 | | Crystal structure of the SVS_A2 protein (205-DREMH-209 /205-AQDLE-209 mutant) from ancestral sequence reconstruction at 2.38 A resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, SVS variant AT2, ... | | Authors: | Rudraraju, R, Schnell, R, Schneider, G. | | Deposit date: | 2019-11-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Engineering of Ancestors as a Tool to Elucidate Structure, Mechanism, and Specificity of Extant Terpene Cyclase.
J.Am.Chem.Soc., 143, 2021
|
|
6TBD
 
 | |
