3LCC
 
 | |
4UMJ
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | | 分子名称: | GERANYLTRANSTRANSFERASE, IBANDRONATE, MAGNESIUM ION | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2014-05-18 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6WCK
 
 | | KRAS G-quadruplex G16T mutant with Bromo Uracil replacing T8 and T16. | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*CP*GP*GP*(BRU)P*GP*TP*GP*GP*GP*AP*AP*(BRU)P*AP*GP*GP*GP*AP*A)-3'), POTASSIUM ION | | 著者 | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | 登録日 | 2020-03-30 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
6N65
 
 | | KRAS G-quadruplex G16T mutant. | | 分子名称: | KRAS G-quadruplex G16T mutant, POTASSIUM ION | | 著者 | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | 登録日 | 2018-11-25 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
7TJF
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA, 84 bp ARS1, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJK
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with docked Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJJ
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with docked Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJI
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 2) with flexible Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
7TJH
 
 | | S. cerevisiae ORC bound to 84 bp ARS1 DNA and Cdc6 (state 1) with flexible Orc6 N-terminal domain | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 6, DNA, ... | | 著者 | Schmidt, J.M, Yang, R, Kumar, A, Hunker, O, Bleichert, F. | | 登録日 | 2022-01-16 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | A mechanism of origin licensing control through autoinhibition of S. cerevisiae ORC·DNA·Cdc6.
Nat Commun, 13, 2022
|
|
3BJX
 
 | |
2NO5
 
 | | Crystal Structure analysis of a Dehalogenase with intermediate complex | | 分子名称: | (2S)-2-CHLOROPROPANOIC ACID, (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, ... | | 著者 | Schmidberger, J.W, Wilce, M.C.J. | | 登録日 | 2006-10-24 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
2NO4
 
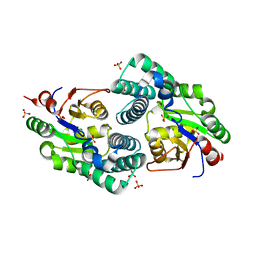 | | Crystal Structure analysis of a Dehalogenase | | 分子名称: | (S)-2-haloacid dehalogenase IVA, CHLORIDE ION, SULFATE ION | | 著者 | Schmidberger, J.W, Wilce, M.C.J. | | 登録日 | 2006-10-24 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Crystal structures of the substrate free-enzyme, and reaction intermediate of the HAD superfamily member, haloacid dehalogenase DehIVa from Burkholderia cepacia MBA4
J.Mol.Biol., 368, 2007
|
|
6EEN
 
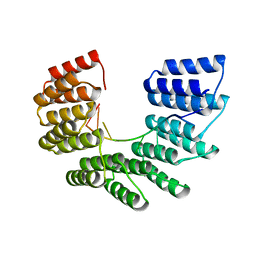 | |
6ZHI
 
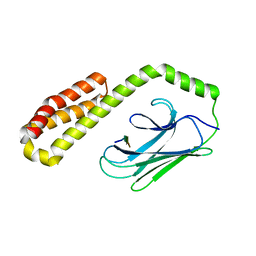 | |
7JGS
 
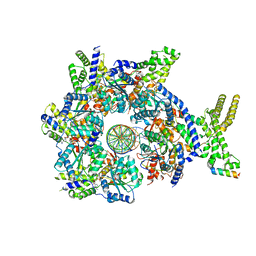 | |
7JK4
 
 | |
7JK6
 
 | | Structure of Drosophila ORC in the active conformation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | 著者 | Schmidt, J.M, Bleichert, F. | | 登録日 | 2020-07-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
7JGR
 
 | |
7JK2
 
 | |
7JK3
 
 | |
7JK5
 
 | | Structure of Drosophila ORC bound to DNA | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA (32-MER), MAGNESIUM ION, ... | | 著者 | Schmidt, J.M, Bleichert, F. | | 登録日 | 2020-07-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6.
Nat Commun, 11, 2020
|
|
4B9E
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa, with bound MFA. | | 分子名称: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION, ... | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2012-09-04 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4B9A
 
 | | Structure of a putative epoxide hydrolase from Pseudomonas aeruginosa. | | 分子名称: | GLYCEROL, PROBABLE EPOXIDE HYDROLASE, SULFATE ION | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2012-09-03 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3ZOU
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | | 分子名称: | 3-(2-oxo-1,3-benzoxazol-3(2H)-yl)propanoic acid, DIMETHYL SULFOXIDE, FARNESYL PYROPHOSPHATE SYNTHASE, ... | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2013-02-25 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3ZL6
 
 | | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PAO1, with bound fragment KM10833. | | 分子名称: | 2-(1,2-benzoxazol-3-yl)ethanoic acid, DIMETHYL SULFOXIDE, GERANYLTRANSTRANSFERASE, ... | | 著者 | Schmidberger, J.W, Schnell, R, Schneider, G. | | 登録日 | 2013-01-28 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Characterization of Substrate and Inhibitor Binding to Farnesyl Pyrophosphate Synthase from Pseudomonas Aeruginosa.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
