1BW5
 
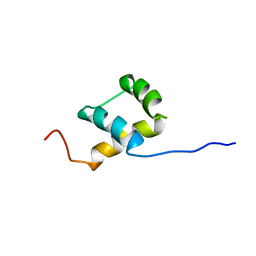 | | THE NMR SOLUTION STRUCTURE OF THE HOMEODOMAIN OF THE RAT INSULIN GENE ENHANCER PROTEIN ISL-1, 50 STRUCTURES | | 分子名称: | INSULIN GENE ENHANCER PROTEIN ISL-1 | | 著者 | Ippel, J.H, Larsson, G, Behravan, G, Zdunek, J, Lundqvist, M, Schleucher, J, Lycksell, P.-O, Wijmenga, S.S. | | 登録日 | 1998-09-29 | | 公開日 | 1999-06-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the homeodomain of the rat insulin-gene enhancer protein isl-1. Comparison with other homeodomains.
J.Mol.Biol., 288, 1999
|
|
1O8T
 
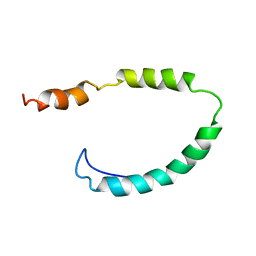 | | Global Structure and Dynamics of Human Apolipoprotein CII in Complex with Micelles: Evidence for increased mobility of the helix involved in the activation of lipoprotein lipase | | 分子名称: | APOLIPOPROTEIN C-II | | 著者 | Zdunek, J, Martinez, G.V, Schleucher, J, Lycksell, P.O, Yin, Y, Nilsson, S, Shen, Y, Olivecrona, G, Wijmenga, S. | | 登録日 | 2002-11-29 | | 公開日 | 2003-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global Structure and Dynamics of Human Apolipoprotein Cii in Complex with Micelles: Evidence for Increased Mobility of the Helix Involved in the Activation of Lipoprotein Lipase
Biochemistry, 42, 2003
|
|
2LVF
 
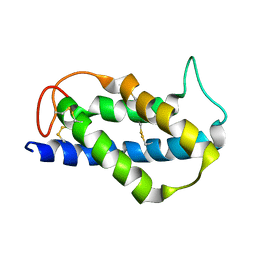 | | Solution structure of the Brazil Nut 2S albumin Ber e 1 | | 分子名称: | 2S albumin | | 著者 | Rundqvist, L, Tengel, T, Zdunek, J, Schleucher, J, Alcocer, M.J, Larsson, G. | | 登録日 | 2012-07-04 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure, copper binding and backbone dynamics of recombinant Ber e 1-the major allergen from Brazil nut.
Plos One, 7, 2012
|
|
2F1Q
 
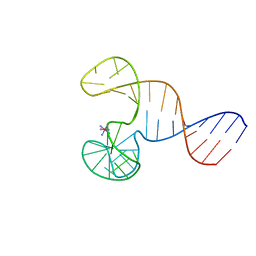 | |
1SNJ
 
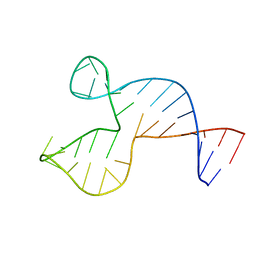 | | Solution structure of the DNA three-way junction with the A/C-stacked conformation | | 分子名称: | 36-MER | | 著者 | Wu, B, Girard, F, van Buuren, B, Schleucher, J, Tessari, M, Wijmenga, S. | | 登録日 | 2004-03-11 | | 公開日 | 2005-04-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Global structure of a DNA three-way junction by solution NMR: towards prediction of 3H fold.
Nucleic Acids Res., 32, 2004
|
|
2IXZ
 
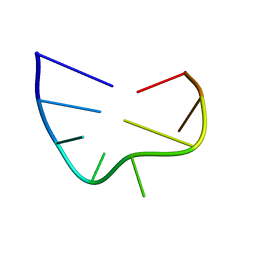 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | 分子名称: | 5'-R(*GP*CP*UP*GP*UP*GP*CP*CP)-3' | | 著者 | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-09-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
2IXY
 
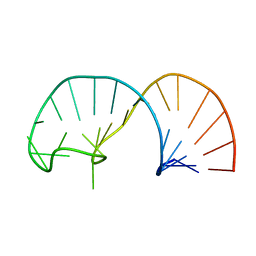 | | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal | | 分子名称: | 5'-R(*GP*GP*CP*CP*UP*CP*CP*AP*AP*GP *CP*UP*GP*UP*GP*CP*CP*UP*UP*GP*GP*GP*UP*GP*GP*CP*C)-3' | | 著者 | Flodell, S, Petersen, M, Girard, F, Zdunek, J, Kidd-Ljunggren, K, Schleucher, J, Wijmenga, S.S. | | 登録日 | 2006-07-11 | | 公開日 | 2006-09-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal.
Nucleic Acids Res., 34, 2006
|
|
