1E8E
 
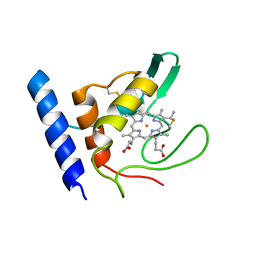 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
1E8J
 
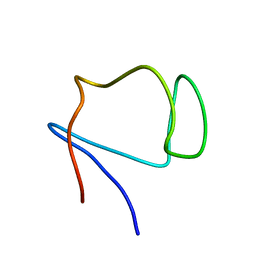 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS ZINC RUBREDOXIN, NMR, 20 STRUCTURES | | Descriptor: | RUBREDOXIN | | Authors: | Lamosa, P, Brennan, L, Vis, H, Turner, D.L, Santos, H. | | Deposit date: | 2000-09-21 | | Release date: | 2001-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Desulfovibrio gigas rubredoxin: a model for studying protein stabilization by compatible solutes.
Extremophiles, 5, 2001
|
|
5AOZ
 
 | | High resolution SeMet structure of the third cohesin from Ruminococcus flavefaciens scaffoldin protein, ScaB | | Descriptor: | GLYCEROL, PUTATIVE CELLULOSOMAL SCAFFOLDIN PROTEIN | | Authors: | Bule, P, Carvalho, A.L, Santos, H, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural Characterization of the Third Cohesin from Ruminococcus Flavefaciens Scaffoldin Protein, Scab
To be Published
|
|
1SPW
 
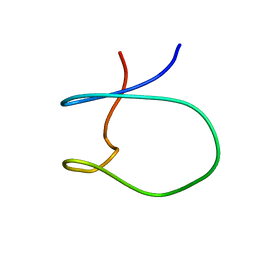 | | Solution Structure of a Loop Truncated Mutant from D. gigas Rubredoxin, NMR | | Descriptor: | Rubredoxin | | Authors: | Pais, T.M, Lamosa, P, dos Santos, W, LeGall, J, Turner, D.L, Santos, H. | | Deposit date: | 2004-03-17 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of protein stabilization by solutes: the importance of the hairpin loop in rubredoxins
FEBS J., 272, 2005
|
|
1OAE
 
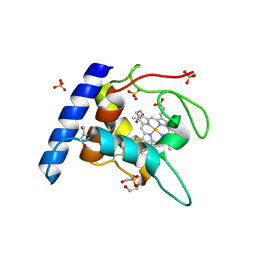 | | Crystal structure of the reduced form of cytochrome c" from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C", GLYCEROL, HEME C, ... | | Authors: | Enguita, F.J, Grenha, R, Santos, H, Carrondo, M.A. | | Deposit date: | 2003-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
4UZN
 
 | | The native structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B) | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
2WVK
 
 | | Mannosyl-3-phosphoglycerate synthase from Thermus thermophilus HB27 apoprotein | | Descriptor: | CITRATE ANION, MANNOSYL-3-PHOSPHOGLYCERATE SYNTHASE, ZINC ION | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Victor, B, Soares, C.M, Santos, H, Matias, P.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Analysis of Thermus Thermophilus Hb27 Mannosyl-3-Phosphoglycerate Synthase Provides Evidence for a Second Catalytic Metal Ion and New Insight Into the Retaining Mechanism of Glycosyltransferases.
J.Biol.Chem., 285, 2010
|
|
2WVM
 
 | | H309A mutant of Mannosyl-3-phosphoglycerate synthase from Thermus thermophilus HB27 in complex with GDP-alpha-D-Mannose and Mg(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE SYNTHASE, ... | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Victor, B, Soares, C.M, Santos, H, Matias, P.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.977 Å) | | Cite: | Structural Analysis of Thermus Thermophilus Hb27 Mannosyl-3-Phosphoglycerate Synthase Provides Evidence for a Second Catalytic Metal Ion and New Insight Into the Retaining Mechanism of Glycosyltransferases.
J.Biol.Chem., 285, 2010
|
|
2XME
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE, GLYCEROL | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Archaeoglobus Fulgidus Ctp:Inositol-1-Phosphate Cytidylyltransferase, a Key Enzyme for Di-Myo-Inositol-Phosphate Synthesis in (Hyper)Thermophiles.
J.Bacteriol., 193, 2011
|
|
2XMH
 
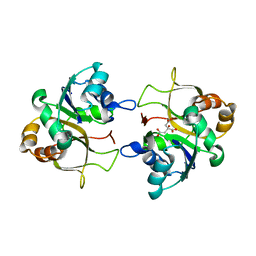 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
2WVL
 
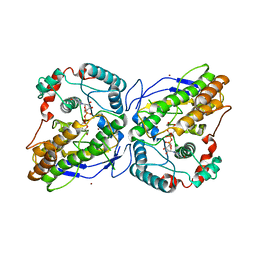 | | Mannosyl-3-phosphoglycerate synthase from Thermus thermophilus HB27 in complex with GDP-alpha-D-Mannose and Mg(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE SYNTHASE, ... | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Victor, B, Soares, C.M, Santos, H, Matias, P.M. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural Analysis of Thermus Thermophilus Hb27 Mannosyl-3-Phosphoglycerate Synthase Provides Evidence for a Second Catalytic Metal Ion and New Insight Into the Retaining Mechanism of Glycosyltransferases.
J.Biol.Chem., 285, 2010
|
|
1QN0
 
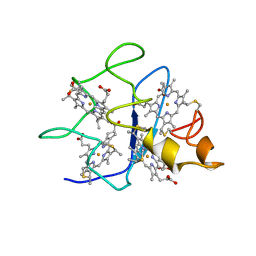 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Teodoro, M.L, Brennan, L, Legall, J, Santos, H, Xavier, A.V, Turner, D.L. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-12 | | Last modified: | 2019-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Network of Functional Cooperativities in Cytochrome C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
5D92
 
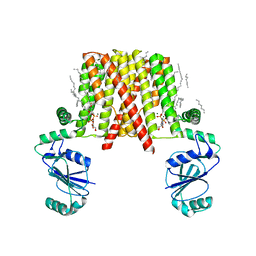 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
1A2I
 
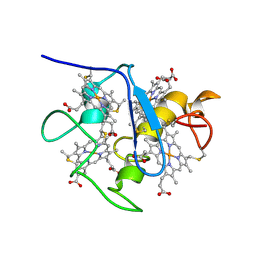 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
5D91
 
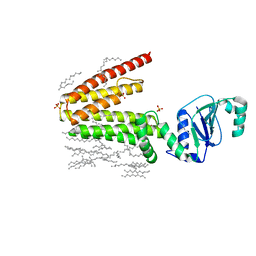 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | Descriptor: | AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, Octadecane, ... | | Authors: | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
1GU2
 
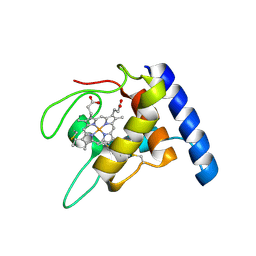 | | Crystal structure of oxidized cytochrome c'' from Methylophilus methylotrophus | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Enguita, F.J, Pohl, E, Rodrigues, A, Santos, H, Carrondo, M.A. | | Deposit date: | 2002-01-22 | | Release date: | 2003-01-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Evidence for a Proton Transfer Pathway Coupled with Haem Reduction of Cytochrome C" from Methylophilus Methylotrophus.
J.Biol.Inorg.Chem., 11, 2006
|
|
4BND
 
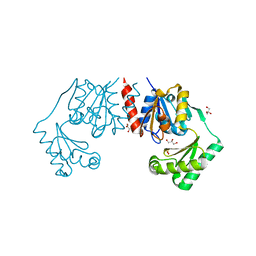 | | Structure of an atypical alpha-phosphoglucomutase similar to eukaryotic phosphomannomutases | | Descriptor: | ALPHA-PHOSPHOGLUCOMUTASE, GLYCEROL, SULFATE ION | | Authors: | Nogly, P, Matias, P.M, De Rosa, M, Castro, R, Santos, H, Neves, A.R, Archer, M. | | Deposit date: | 2013-05-14 | | Release date: | 2013-10-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-Resolution Structure of an Atypical [Alpha]-Phosphoglucomutase Related to Eukaryotic Phosphomannomutases
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MND
 
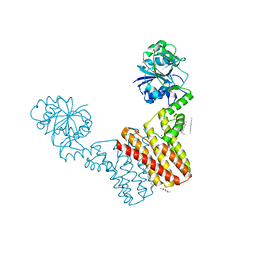 | | Crystal structure of Archaeoglobus fulgidus IPCT-DIPPS bifunctional membrane protein | | Descriptor: | CTP L-myo-inositol-1-phosphate cytidylyltransferase/CDP-L-myo-inositol myo-inositolphosphotransferase, EICOSANE, MAGNESIUM ION | | Authors: | Nogly, P, Gushchin, I, Remeeva, A, Esteves, A.M, Ishchenko, A, Ma, P, Grudinin, S, Borges, N, Round, E, Moraes, I, Borshchevskiy, V, Santos, H, Gordeliy, V, Archer, M. | | Deposit date: | 2013-09-10 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-ray structure of a CDP-alcohol phosphatidyltransferase membrane enzyme and insights into its catalytic mechanism.
Nat Commun, 5, 2014
|
|
6WM5
 
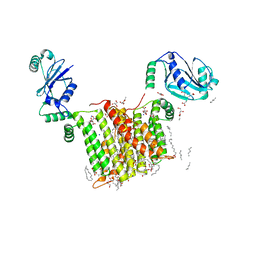 | | Structure of a phosphatidylinositol-phosphate synthase (PIPS) from Mycobacterium kansasii | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, 3,3',3''-phosphanetriyltripropanoic acid, ... | | Authors: | Belcher Dufrisne, M, Jorge, C.D, Timoteo, C.G, Petrou, V.I, Ashraf, K.U, Banerjee, S, Clarke, O.B, Santos, H, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-04-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Structural and Functional Characterization of Phosphatidylinositol-Phosphate Biosynthesis in Mycobacteria.
J.Mol.Biol., 432, 2020
|
|
4V2X
 
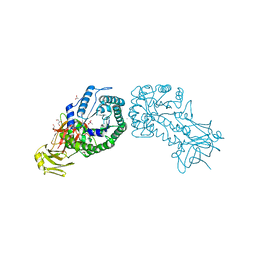 | | High resolution structure of the full length tri-modular endo-beta-1, 4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
4UZ8
 
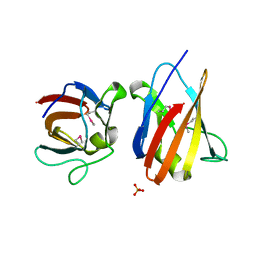 | | The SeMet structure of the family 46 carbohydrate-binding module (CBM46) of endo-beta-1,4-glucanase B (Cel5B) from Bacillus halodurans | | Descriptor: | ENDO-BETA-1,4-GLUCANASE (CELULASE B), SULFATE ION | | Authors: | Venditto, I, Santos, H, Ferreira, L.M.A, Sakka, K, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family 46 Carbohydrate-Binding Modules Contribute to the Enzymatic Hydrolysis of Xyloglucan and Beta-1,3-1,4-Glucans Through Distinct Mechanisms.
J.Biol.Chem., 290, 2015
|
|
3ZWK
 
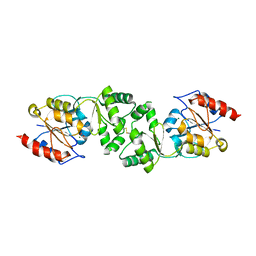 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the metavanadate | | Descriptor: | MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, VANADATE ION | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-08-01 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZTW
 
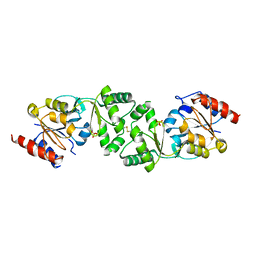 | | The 3-dimensional structure of apo-MpgP, the mannosyl-3- phosphoglycerate phosphatase from Thermus thermophilus HB27 in its apo-form | | Descriptor: | MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, PHOSPHATE ION | | Authors: | Goncalves, S, Borges, N, Esteves, A.M, Santos, H, Matias, P.M. | | Deposit date: | 2011-07-12 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZUP
 
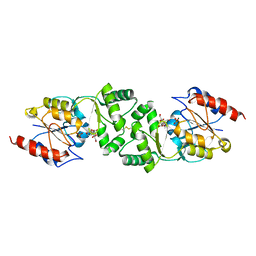 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate and orthophosphate reaction products. | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE, ... | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
3ZWD
 
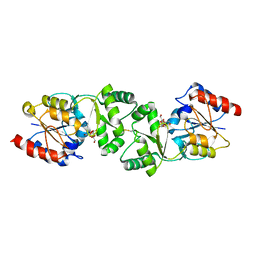 | | The 3-dimensional structure of MpgP from Thermus thermophilus HB27, in complex with the alpha-mannosylglycerate. | | Descriptor: | (2R)-3-hydroxy-2-(alpha-D-mannopyranosyloxy)propanoic acid, MAGNESIUM ION, MANNOSYL-3-PHOSPHOGLYCERATE PHOSPHATASE | | Authors: | Goncalves, S, Esteves, A.M, Santos, H, Borges, N, Matias, P.M. | | Deposit date: | 2011-07-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | The Three-Dimensional Structure of Mannosyl-3-Phosphoglycerate Phosphatase from Thermus Thermophilus Hb27: A New Member of the Haloalkanoic Acid Dehalogenase Superfamily.
Biochemistry, 50, 2011
|
|
