3E53
 
 | |
4RRR
 
 | |
3H2H
 
 | |
3H2I
 
 | |
3H2G
 
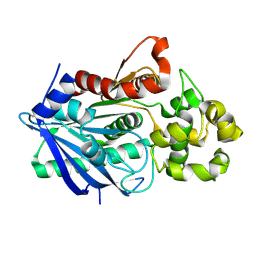 | |
3H2J
 
 | |
3H2K
 
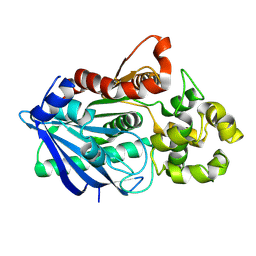
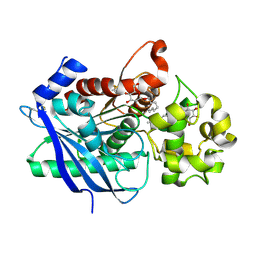 | | Crystal structure of a ligand-bound form of the rice cell wall degrading esterase LipA from Xanthomonas oryzae | | Descriptor: | esterase, octyl beta-D-glucopyranoside | | Authors: | Aparna, G, Chatterjee, A, Sonti, R.V, Sankaranarayanan, R. | | Deposit date: | 2009-04-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Cell Wall-Degrading Esterase of Xanthomonas oryzae Requires a Unique Substrate Recognition Module for Pathogenesis on Rice
Plant Cell, 21, 2009
|
|
1T2N
 
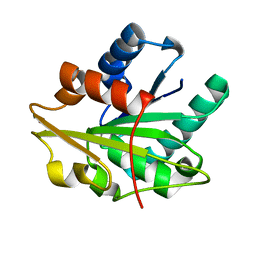 | |
1T4M
 
 | |
3I9H
 
 | |
1KOG
 
 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
3HZB
 
 | |
3T5B
 
 | |
3T5A
 
 | |
3T5C
 
 | |
3ENT
 
 | |
3ENU
 
 | |
3CW3
 
 | | Crystal structure of AIM1g1 | | Descriptor: | Absent in melanoma 1 protein, GLYCEROL | | Authors: | Aravind, P, Sankaranarayanan, R, Sharma, Y. | | Deposit date: | 2008-04-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Exploring the limits of sequence and structure in a variant betagamma-crystallin domain of the protein absent in melanoma-1 (AIM1).
J.Mol.Biol., 381, 2008
|
|
1LJ3
 
 | |
1LJK
 
 | |
1LJ4
 
 | |
1LJI
 
 | |
1LJE
 
 | |
1LJJ
 
 | |
1LJF
 
 | |
