4II2
 
 | | Crystal structure of Ubiquitin activating enzyme 1 (Uba1) in complex with the Ub E2 Ubc4, ubiquitin, and ATP/Mg | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Olsen, S.K, Lima, C.D. | | Deposit date: | 2012-12-19 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer.
Mol.Cell, 49, 2013
|
|
4IKH
 
 | | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutathione S-transferase | | Authors: | Vetting, M.W, Sauder, J.M, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Burley, S.K, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-26 | | Release date: | 2013-01-16 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a glutathione transferase family member from Pseudomonas fluorescens pf-5, target efi-900003, with two glutathione bound
To be Published
|
|
1JSY
 
 | | Crystal structure of bovine arrestin-2 | | Descriptor: | Bovine arrestin-2 (full length) | | Authors: | Milano, S.K, Pace, H.C, Kim, Y.M, Brenner, C, Benovic, J.L. | | Deposit date: | 2001-08-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis.
Biochemistry, 41, 2002
|
|
4JR8
 
 | |
1JME
 
 | | Crystal Structure of Phe393His Cytochrome P450 BM3 | | Descriptor: | BIFUNCTIONAL P-450:NADPH-P450 REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ost, T.W.B, Munro, A.W, Mowat, C.G, Pesseguiero, A, Fulco, A.J, Cho, A.K, Cheesman, M.A, Walkinshaw, M.D, Chapman, S.K. | | Deposit date: | 2001-07-18 | | Release date: | 2001-11-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and spectroscopic analysis of the F393H mutant of flavocytochrome P450 BM3.
Biochemistry, 40, 2001
|
|
1L0C
 
 | | Investigation of the Roles of Catalytic Residues in Serotonin N-Acetyltransferase | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Scheibner, K.A, De Angelis, J, Burley, S.K, Cole, P.A. | | Deposit date: | 2002-02-08 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigation of the roles of catalytic residues in serotonin N-acetyltransferase.
J.Biol.Chem., 277, 2002
|
|
7M0O
 
 | | DGT-28 EPSPS | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Characterization of a Glyphosate-Tolerant Enzyme from Streptomyces svecius : A Distinct Class of 5-Enolpyruvylshikimate-3-phosphate Synthases.
J.Agric.Food Chem., 69, 2021
|
|
1KCX
 
 | | X-ray structure of NYSGRC target T-45 | | Descriptor: | DIHYDROPYRIMIDINASE RELATED PROTEIN-1 | | Authors: | Deo, R.C, Schmidt, E.F, Strittmatter, S.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-11-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural bases for CRMP function in plexin-dependent semaphorin3A signaling
Embo J., 23, 2004
|
|
4GS1
 
 | | Crystal structure of DyP-type peroxidase from Thermobifida cellulosilytica | | Descriptor: | DyP-type peroxidase, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-27 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
7MSK
 
 | | ThuS glycosin S-glycosyltransferase | | Descriptor: | Glyco_trans_2-like domain-containing protein, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSN
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
7MSP
 
 | | SunS glycosin S-glycosyltransferase | | Descriptor: | MAGNESIUM ION, SPbeta prophage-derived glycosyltransferase SunS, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Garg, N, Nair, S.K. | | Deposit date: | 2021-05-11 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic investigations of protein S-glycosyltransferases.
Cell Chem Biol, 28, 2021
|
|
1JOY
 
 | | SOLUTION STRUCTURE OF THE HOMODIMERIC DOMAIN OF ENVZ FROM ESCHERICHIA COLI BY MULTI-DIMENSIONAL NMR. | | Descriptor: | PROTEIN (ENVZ_ECOLI) | | Authors: | Tomomori, C, Tanaka, T, Dutta, R, Park, H, Saha, S.K, Zhu, Y, Ishima, R, Liu, D, Tong, K.I, Kurokawa, H, Qian, H, Inouye, M, Ikura, M. | | Deposit date: | 1998-12-28 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ.
Nat.Struct.Biol., 6, 1999
|
|
1LW4
 
 | |
1M64
 
 | | Crystal structure of Q363F mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mowat, C.G, Pankhurst, K.L, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-07-12 | | Release date: | 2002-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering water to act as an active site acid catalyst in a soluble fumarate reductase
Biochemistry, 41, 2002
|
|
4GU7
 
 | | Crystal structure of DyP-type peroxidase (SCO7193) from Streptomyces coelicolor | | Descriptor: | NICKEL (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Putative uncharacterized protein SCO7193 | | Authors: | Lukk, T, Hetta, A.M.A, Jones, A, Solbiati, J, Majumdar, S, Cronan, J.E, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2012-08-29 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | DyP-type peroxidases from Stretptomyces and Thermobifida can modify organosolv lignin.
To be Published
|
|
4H6W
 
 | |
4HCQ
 
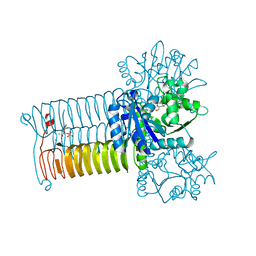 | | Crystal structure of GLMU from mycobacterium tuberculosis in complex with glucosamine-1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein GlmU, COBALT (II) ION, ... | | Authors: | Jagtap, P.K.A, Verma, S.K, Vithani, N. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures identify an atypical two-metal-ion mechanism for uridyltransfer in GlmU: its significance to sugar nucleotidyl transferases
J.Mol.Biol., 425, 2013
|
|
4K3C
 
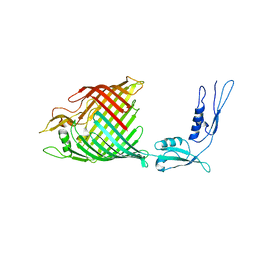 | | The crystal structure of BamA from Haemophilus ducreyi lacking POTRA domains 1-3 | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
4K3B
 
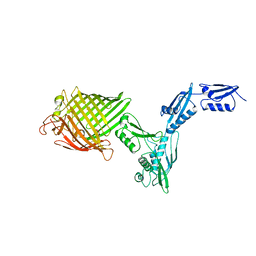 | | The crystal structure of BamA from Neisseria gonorrhoeae | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Noinaj, N, Lukacik, P, Chang, H, Easley, N, Buchanan, S.K. | | Deposit date: | 2013-04-10 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insight into the biogenesis of beta-barrel membrane proteins.
Nature, 501, 2013
|
|
4K5R
 
 | | The 2.0 angstrom crystal structure of MTMOIV, a baeyer-villiger monooxygenase from the mithramycin biosynthetic pathway in streptomyces argillaceus. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Bosserman, M.A, Rohr, J, Buchanan, S.K. | | Deposit date: | 2013-04-15 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Insight into Substrate Recognition and Catalysis of Baeyer-Villiger Monooxygenase MtmOIV, the Key Frame-Modifying Enzyme in the Biosynthesis of Anticancer Agent Mithramycin.
Acs Chem.Biol., 8, 2013
|
|
1LJ1
 
 | | Crystal structure of Q363F/R402A mutant flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mowat, C.G, Pankhurst, K.L, Miles, C.S, Leys, D, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2002-04-18 | | Release date: | 2002-11-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering Water to act as the active site acid catalyst in a soluble fumarate reductase
Biochemistry, 41, 2002
|
|
1MBU
 
 | | Crystal Structure Analysis of ClpSN heterodimer | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, BIS-(2-HYDROXYETHYL)AMINO-TRIS(HYDROXYMETHYL)METHANE YTTRIUM, CHLORIDE ION, ... | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of the AAA+ Chaperone, ClpA
J.Biol.Chem., 277, 2002
|
|
1LW7
 
 | | NADR PROTEIN FROM HAEMOPHILUS INFLUENZAE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, TRANSCRIPTIONAL REGULATOR NADR | | Authors: | Singh, S.K, Kurnasov, O.V, Chen, B, Robinson, H, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-05-30 | | Release date: | 2002-08-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities.
J.Biol.Chem., 277, 2002
|
|
4HYR
 
 | | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hegde, R.P, Toro, R, Burley, S.K, Almo, S.C, Ramagopal, U.A, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure of putative Glucarate dehydratase from Acidaminococcus sp. D21 with unusual static disorder
To be published
|
|
