1PQ7
 
 | | Trypsin at 0.8 A, pH5 / borax | | Descriptor: | ARGININE, SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQA
 
 | | Trypsin with PMSF at atomic resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PPZ
 
 | | Trypsin complexes at atomic and ultra-high resolution | | Descriptor: | SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQ8
 
 | | Trypsin at pH 4 at atomic resolution | | Descriptor: | CITRIC ACID, GLY-GLY-ARG PEPTIDE, LYSINE, ... | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
1PQ5
 
 | | Trypsin at pH 5, 0.85 A | | Descriptor: | ARGININE, SULFATE ION, Trypsin | | Authors: | Schmidt, A, Jelsch, C, Rypniewski, W, Lamzin, V.S. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Trypsin Revisited: CRYSTALLOGRAPHY AT (SUB) ATOMIC RESOLUTION AND QUANTUM CHEMISTRY REVEALING DETAILS OF CATALYSIS.
J.Biol.Chem., 278, 2003
|
|
3KI8
 
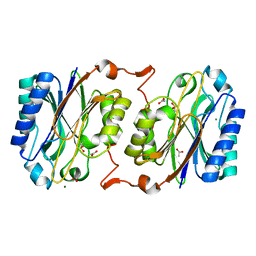 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, Beta ureidopropionase (Beta-alanine synthase), MAGNESIUM ION | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-10-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
3KLC
 
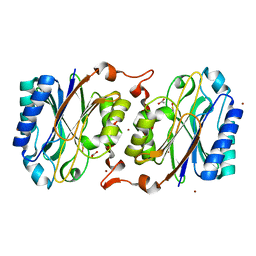 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, BROMIDE ION, Beta ureidopropionase (Beta-alanine synthase), ... | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
3GM7
 
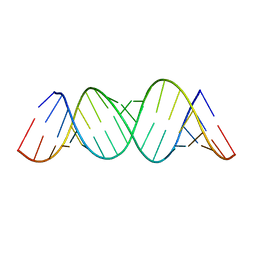 | |
3IVZ
 
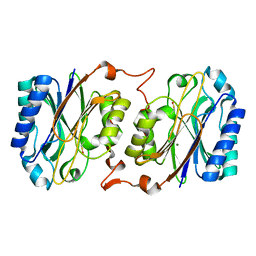 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | MAGNESIUM ION, Nitrilase | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
3IW3
 
 | | Crystal structure of hyperthermophilic nitrilase | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Nitrilase | | Authors: | Raczynska, J, Vorgias, C, Antranikian, G, Rypniewski, W. | | Deposit date: | 2009-09-02 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of a thermoactive nitrilase.
J.Struct.Biol., 173, 2010
|
|
3JXR
 
 | |
3NJ7
 
 | | 1.9 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
3O5W
 
 | | Binding of kinetin in the active site of mistletoe lectin I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Malecki, P.H, Meyer, A, Rypniewski, W, Szymanski, M, Barciszewski, J, Betzel, C. | | Deposit date: | 2010-07-28 | | Release date: | 2011-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding of the plant hormone kinetin in the active site of Mistletoe Lectin I from Viscum album.
Biochim.Biophys.Acta, 1824, 2012
|
|
3O8N
 
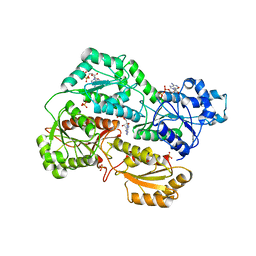 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3NJ6
 
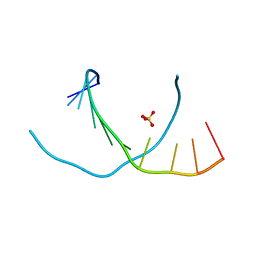 | | 0.95 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
3GLP
 
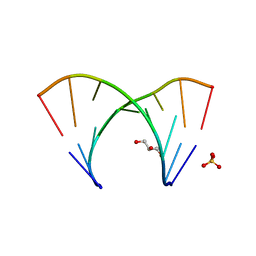 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
3O8O
 
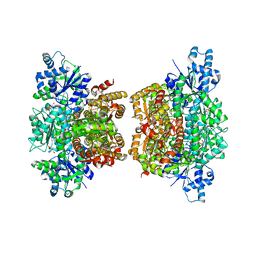 | | Structure of phosphofructokinase from Saccharomyces cerevisiae | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase subunit alpha, ... | | Authors: | Banaszak, K, Mechin, I, Kopperschlager, G, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3JXQ
 
 | | X-Ray structure of r[CGCG(5-fluoro)CG]2 | | Descriptor: | MAGNESIUM ION, r[CGCG(5-fluoro)CG]2 | | Authors: | Adamiak, D.A, Milecki, J, Adamiak, R.W, Rypniewski, W. | | Deposit date: | 2009-09-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The hydration and unusual hydrogen bonding in the crystal structure of an RNA duplex containing alternating CG base pairs
New J.Chem., 34, 2010
|
|
1Q5T
 
 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1R3O
 
 | | Crystal structure of the first RNA duplex in L-conformation at 1.9A resolution | | Descriptor: | L-RNA | | Authors: | Vallazza, M, Perbandt, M, Klussmann, S, Rypniewski, W, Erdmann, V.A, Betzel, C. | | Deposit date: | 2003-10-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First look at RNA in L-configuration.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TJD
 
 | | The crystal structure of the reduced disulphide bond isomerase, DsbC, from Escherichia coli | | Descriptor: | Thiol:disulfide interchange protein dsbC | | Authors: | Banaszak, K, Mechin, I, Frost, G, Rypniewski, W. | | Deposit date: | 2004-06-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the reduced disulfide-bond isomerase DsbC from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4E59
 
 | |
4HMD
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction intermediate - oxazolinium ion (NGO) | | Descriptor: | 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HME
 
 | | Crystal structure of cold-adapted chitinase from Moritella marina with a reaction product - NAG2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase 60, GLYCEROL, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Raczynska, J.E, Rypniewski, W. | | Deposit date: | 2012-10-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of a complete four-domain chitinase from Moritella marina, a marine psychrophilic bacterium
Acta Crystallogr.,Sect.D, 69, 2013
|
|
