6SVF
 
 | | Crystal structure of the P235GK mutant of ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Vitagliano, L, Berisio, R, Esposito, L, Balasco, N, Smaldone, G, Ruggiero, A. | | Deposit date: | 2019-09-18 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The non-swapped monomeric structure of the arginine-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
2Z53
 
 | |
4KPM
 
 | | Crystal structure of the catalytic domain of RpfB from Mycobacterium tuberculosis in complex with triNAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, Resuscitation-promoting factor RpfB, ... | | Authors: | Squeglia, F, Ruggiero, A, Berisio, R. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Carbohydrate Recognition by RpfB from Mycobacterium tuberculosis Unveiled by Crystallographic and Molecular Dynamics Analyses.
Biophys.J., 104, 2013
|
|
4KL7
 
 | | Crystal structure of the catalytic domain of RpfB from Mycobacterium tuberculosis | | Descriptor: | Resuscitation-promoting factor RpfB, SULFATE ION | | Authors: | Squeglia, F, Romano, M, Ruggiero, A, Berisio, R. | | Deposit date: | 2013-05-07 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Carbohydrate Recognition by RpfB from Mycobacterium tuberculosis Unveiled by Crystallographic and Molecular Dynamics Analyses.
Biophys.J., 104, 2013
|
|
6QMN
 
 | | Crystal structure of a Ribonuclease A-Onconase chimera | | Descriptor: | PHOSPHATE ION, Ribonuclease pancreatic | | Authors: | Esposito, L, Vitagliano, L, Ruggiero, A, Picone, D, Leone, S, Donnarumma, F. | | Deposit date: | 2019-02-07 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure, stability and aggregation propensity of a Ribonuclease A-Onconase chimera.
Int.J.Biol.Macromol., 133, 2019
|
|
4JMX
 
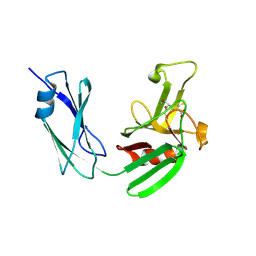 | | Structure of LD transpeptidase LdtMt1 in complex with imipenem | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Probable L,D-transpeptidase LdtA | | Authors: | Correale, S, Ruggiero, A, Capparelli, R, Pedone, E, Berisio, R. | | Deposit date: | 2013-03-14 | | Release date: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of free and inhibited forms of the L,D-transpeptidase LdtMt1 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2MGV
 
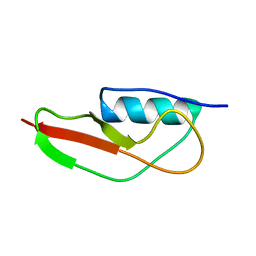 | | NMR structure of PASTA domain of PonA2 from Mycobacterium tuberculosis | | Descriptor: | Bifunctional membrane-associated penicillin-binding protein 1A/1B ponA2 | | Authors: | Calvanese, L, Falcigno, L, Maglione, C, Marasco, D, Ruggiero, A, Squeglia, F, Berisio, R, D'Auria, G. | | Deposit date: | 2013-11-11 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and binding properties of the PASTA domain of PonA2, a key penicillin binding protein from Mycobacterium tuberculosis.
Biopolymers, 101, 2014
|
|
4Q4N
 
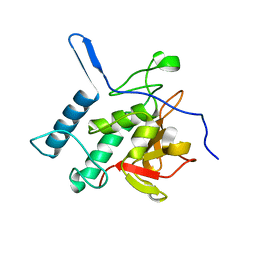 | | Structure of the Resuscitation Promoting Factor Interacting protein RipA mutated at H432 | | Descriptor: | Peptidoglycan endopeptidase RipA | | Authors: | Squeglia, F, Ruggiero, A, Romano, M, Vitagliano, L, Berisio, R. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: evidence for the L-to-D inversion of configuration of the catalytic cysteine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q4T
 
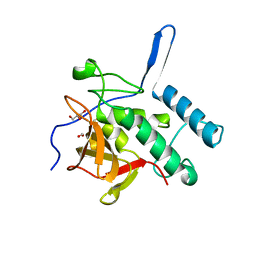 | | Structure of the Resuscitation Promoting Factor Interacting protein RipA mutated at E444 | | Descriptor: | FORMIC ACID, GLYCEROL, Peptidoglycan endopeptidase RipA | | Authors: | Squeglia, F, Ruggiero, A, Romano, M, Vitagliano, L, Berisio, R. | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: evidence for the L-to-D inversion of configuration of the catalytic cysteine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6GPD
 
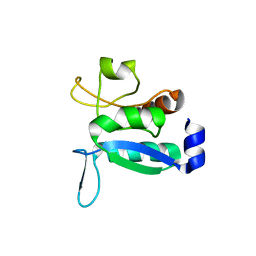 | | Crystal structure of the ligand-free form of domain 1 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
6GGP
 
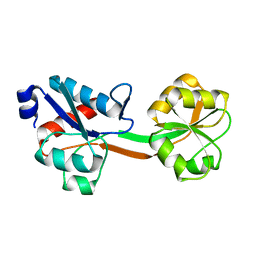 | | Structure of the ligand-free form of truncated ArgBP (residues 20-233) from T. maritima | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-03 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
6GGV
 
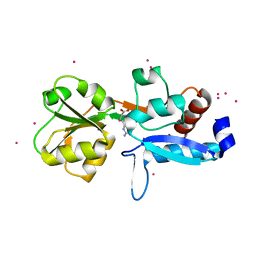 | | Structure of the arginine-bound form of truncated (residues 20-233) ArgBP from T. maritima | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
6GPC
 
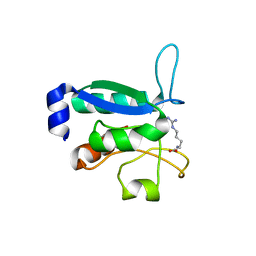 | | Crystal structure of the arginine-bound form of domain 1 from TmArgBP | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
6GPM
 
 | | Crystal structure of domain 2 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-06 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
