5SFQ
 
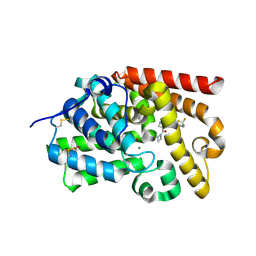 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-(azetidine-1-carbonyl)-N-[1-(2,2-difluoroethyl)pyrazol-3-yl]-2-methylpyrazole-3-carboxamide | | Descriptor: | 4-(azetidine-1-carbonyl)-N-[1-(2,2-difluoroethyl)-1H-pyrazol-3-yl]-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFT
 
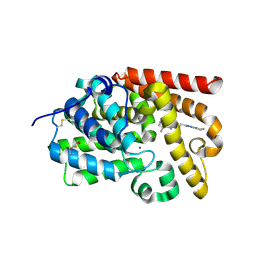 | | Crystal Structure of human phosphodiesterase 10 in complex with 5,8-dimethyl-2-[2-(1-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-a]pyrazine | | Descriptor: | (4S)-5,8-dimethyl-2-{2-[1-methyl-5-(pyrrolidin-1-yl)-1H-1,2,4-triazol-3-yl]ethyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFX
 
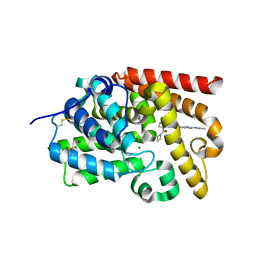 | | Crystal Structure of human phosphodiesterase 10 in complex with N-(2-acetamido-1,3-benzothiazol-5-yl)-4-(azetidine-1-carbonyl)-2-methylpyrazole-3-carboxamide | | Descriptor: | MAGNESIUM ION, N-(2-acetamido-1,3-benzothiazol-5-yl)-4-(azetidine-1-carbonyl)-1-methyl-1H-pyrazole-5-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFY
 
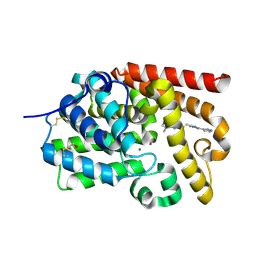 | | Crystal Structure of human phosphodiesterase 10 in complex with 5,8-dimethyl-2-[(E)-2-[2-methyl-5-(1H-pyrazol-4-yl)-1,2,4-triazol-3-yl]ethenyl]-[1,2,4]triazolo[1,5-a]pyrazine | | Descriptor: | (4S)-5,8-dimethyl-2-{(E)-2-[1-methyl-3-(1H-pyrazol-4-yl)-1H-1,2,4-triazol-5-yl]ethenyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SDW
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-N,4-N,2-trimethyl-3-N-(2-phenyl-1H-benzimidazol-5-yl)pyrazole-3,4-dicarboxamide | | Descriptor: | MAGNESIUM ION, N~4~,N~4~,1-trimethyl-N~5~-(2-phenyl-1H-benzimidazol-5-yl)-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEC
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2-[2-(1,4-diphenylimidazol-2-yl)ethyl]-3-methylquinoxaline | | Descriptor: | 2-[2-(1,4-diphenyl-1H-imidazol-2-yl)ethyl]-3-methylquinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SDU
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 5-methyl-4-[2-(1-methyl-4-phenylimidazol-2-yl)ethyl]-2-phenyl-1,3-oxazole | | Descriptor: | 5-methyl-4-[2-(1-methyl-4-phenyl-1H-imidazol-2-yl)ethyl]-2-phenyl-1,3-oxazole, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF4
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-cyclopropyl-N-[2-[2-(dimethylamino)ethyl]-5-pyridin-2-ylpyrazol-3-yl]-3-(pyrimidin-5-ylamino)pyridine-2-carboxamide | | Descriptor: | 6-cyclopropyl-N-{1-[2-(dimethylamino)ethyl]-3-(pyridin-2-yl)-1H-pyrazol-5-yl}-3-[(pyrimidin-5-yl)amino]pyridine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SDV
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2-methyl-4-N-(1,3-oxazol-4-ylmethyl)-3-N-(2-phenylimidazo[1,2-a]pyrimidin-7-yl)pyrazole-3,4-dicarboxamide | | Descriptor: | 1-methyl-N~4~-[(1,3-oxazol-4-yl)methyl]-N~5~-[(4R)-2-phenylimidazo[1,2-a]pyrimidin-7-yl]-1H-pyrazole-4,5-dicarboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEE
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-cyclopropyl-N-(2-pyridin-2-ylpyrazol-3-yl)-3-(pyrimidin-5-ylamino)pyridine-2-carboxamide | | Descriptor: | 6-cyclopropyl-N-[1-(pyridin-2-yl)-1H-pyrazol-5-yl]-3-[(pyrimidin-5-yl)amino]pyridine-2-carboxamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rodriguez-Sarmiento, R.M, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SER
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-N-(2-fluoroethyl)-4-N,2-dimethyl-3-N-(2-phenyl-[1,2,4]triazolo[1,5-a]pyridin-7-yl)pyrazole-3,4-dicarboxamide | | Descriptor: | MAGNESIUM ION, N~4~-(2-fluoroethyl)-N~4~,1-dimethyl-N~5~-[(4S)-2-phenyl[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SF9
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 2-methyl-3-[(1-methyl-4-phenylimidazol-2-yl)methoxy]quinoxaline | | Descriptor: | 2-methyl-3-{[(4R)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]methoxy}quinoxaline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFP
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with N-(2-chloropyridin-4-yl)-2-phenylpyrazolo[1,5-a]pyridine-6-carboxamide | | Descriptor: | (8S)-N-(2-chloropyridin-4-yl)-2-phenylpyrazolo[1,5-a]pyridine-6-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SE0
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-cyclopropyl-N-[2-(2-hydroxyethyl)-5-pyridin-2-ylpyrazol-3-yl]-3-(pyrimidin-5-ylamino)pyridine-2-carboxamide | | Descriptor: | 6-cyclopropyl-N-[1-(2-hydroxyethyl)-3-(pyridin-2-yl)-1H-pyrazol-5-yl]-3-[(pyrimidin-5-yl)amino]pyridine-2-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEH
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-(azetidine-1-carbonyl)-2-methyl-N-(2-morpholin-4-yl-[1,2,4]triazolo[1,5-a]pyridin-7-yl)pyrazole-3-carboxamide | | Descriptor: | 4-(azetidine-1-carbonyl)-1-methyl-N-[(4S)-2-(morpholin-4-yl)[1,2,4]triazolo[1,5-a]pyridin-7-yl]-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Flohr, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEX
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with N-methyl-6-[(6-methylpyridine-2-carbonyl)amino]-2-phenyl-3H-benzimidazole-5-carboxamide | | Descriptor: | MAGNESIUM ION, N-methyl-6-[(6-methylpyridine-2-carbonyl)amino]-2-phenyl-1H-benzimidazole-5-carboxamide, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFF
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with [2-methyl-6-[2-(1-methyl-4-phenylimidazol-2-yl)ethynyl]imidazo[1,2-b]pyridazin-3-yl]methanol | | Descriptor: | MAGNESIUM ION, ZINC ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFV
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-(azetidine-1-carbonyl)-N-(1-cyclopentylpyrazol-3-yl)-2-methylpyrazole-3-carboxamide | | Descriptor: | 4-(azetidine-1-carbonyl)-N-(1-cyclopentyl-1H-pyrazol-3-yl)-1-methyl-1H-pyrazole-5-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Peters, J.U, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SDX
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-[6-[2-(1-methyl-4-phenylimidazol-2-yl)ethyl]pyridin-2-yl]morpholine | | Descriptor: | 4-(6-{2-[(4S)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]ethyl}pyridin-2-yl)morpholine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Groebke-Zbinden, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEG
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 6-[(5-bromo-1-methyl-2-oxopyridine-3-carbonyl)amino]-N-(oxetan-3-yl)-2-phenyl-3H-benzimidazole-5-carboxamide | | Descriptor: | 5-[(5-bromo-1-methyl-2-oxo-1,2-dihydropyridine-3-carbonyl)amino]-N-(oxetan-3-yl)-2-phenyl-1H-benzimidazole-6-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Bleicher, K, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SFR
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 4-N-ethyl-3-N-[2-[3-(2-fluoroethoxy)phenyl]imidazo[1,2-a]pyrimidin-7-yl]-4-N,2-dimethylpyrazole-3,4-dicarboxamide | | Descriptor: | MAGNESIUM ION, N~4~-ethyl-N~5~-{(4S)-2-[3-(2-fluoroethoxy)phenyl]imidazo[1,2-a]pyrimidin-7-yl}-N~4~,1-dimethyl-1H-pyrazole-4,5-dicarboxamide, ZINC ION, ... | | Authors: | Joseph, C, Gobbi, L, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7FYJ
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FYK
 
 | | Crystal Structure of human FABP4 in complex with 5-phenylmethoxy-4-prop-2-enyl-1,2,4-triazole-3-thiol | | Descriptor: | 5-(benzyloxy)-4-(prop-2-en-1-yl)-4H-1,2,4-triazole-3-thiol, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7G06
 
 | | Crystal Structure of human FABP4 in complex with 5-(chloromethyl)-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-5-(chloromethyl)-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FYT
 
 | | Crystal Structure of human FABP4 in complex with 5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-5-[(3-chloro-2-methylphenoxy)methyl]-2-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ceccarelli-Simona, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
