7KR2
 
 | |
7MPE
 
 | |
7MD2
 
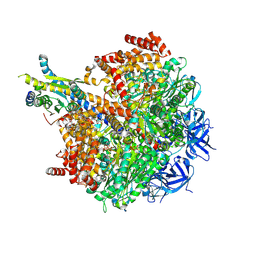 | | The F1 region of ammocidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{Z},7~{E},9~{R},10~{S},11~{E},13~{E},15~{E},17~{R},18~{S},20~{S})-20-[(1~{R})-1-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-[(2~{S},4~{S},5~{S},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-4-oxidanyl-oxan-2-yl]oxy-3-methoxy-6-(3-methoxypropyl)-5-methyl-2,4-bis(oxidanyl)oxan-2-yl]ethyl]-5,18-dimethoxy-3,7,9,11,13,15-hexamethyl-10-[(2~{R},3~{S},4~{R},5~{R},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13,15-hexaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
7MD3
 
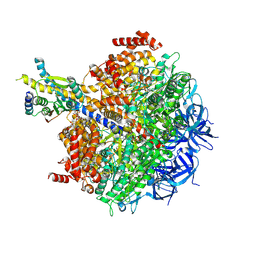 | | The F1 region of apoptolidin-bound Saccharomyces cerevisiae ATP synthase | | Descriptor: | (3~{E},5~{E},7~{E},9~{R},10~{R},11~{E},13~{E},17~{S},18~{S},20~{S})-18-methoxy-20-[(~{R})-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(2~{R})-3-methoxy-2-[(2~{R},4~{S},5~{S},6~{S})-5-[(2~{S},4~{R},5~{R},6~{R})-4-methoxy-6-methyl-5-oxidanyl-oxan-2-yl]oxy-4,6-dimethyl-4-oxidanyl-oxan-2-yl]oxy-propyl]-3,5-dimethyl-2,4-bis(oxidanyl)oxan-2-yl]-oxidanyl-methyl]-10-[(2~{R},3~{S},4~{S},5~{R},6~{S})-5-methoxy-6-methyl-3,4-bis(oxidanyl)oxan-2-yl]oxy-3,5,7,9,13-pentamethyl-17-oxidanyl-1-oxacycloicosa-3,5,7,11,13-pentaen-2-one, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2021-04-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A Family of Glycosylated Macrolides Selectively Target Energetic Vulnerabilities in Leukemia
Biorxiv, 2021
|
|
8SQL
 
 | |
8SQM
 
 | |
8SQ0
 
 | | Cleaved Ycf1p Dimer in the IFwide-beta conformation | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Metal resistance protein YCF1, ... | | Authors: | Bickers, S.C, Benlekbir, S, Rubinstein, J.L, Kanelis, V. | | Deposit date: | 2023-05-04 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a dimeric ABC transporter
To Be Published
|
|
8UX1
 
 | |
3J0J
 
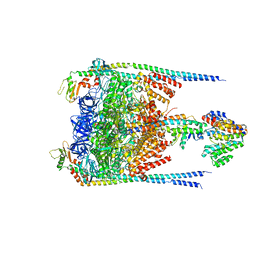 | |
3J9T
 
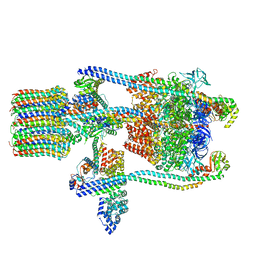 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9U
 
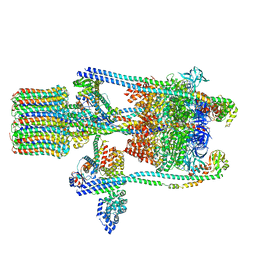 | | Yeast V-ATPase state 2 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9V
 
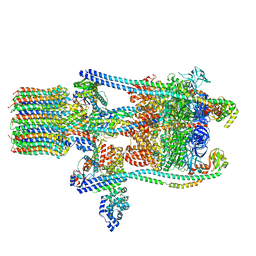 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
6N30
 
 | | Bacillus PS3 ATP synthase class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Y
 
 | | Bacillus PS3 ATP synthase class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2D
 
 | | Bacillus PS3 ATP synthase membrane region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Z
 
 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6O7X
 
 | | Saccharomyces cerevisiae V-ATPase Stv1-V1VO State 3 | | Descriptor: | Putative protein YPR170W-B, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Vasanthakumar, T, Bueler, S.A, Wu, D, Beilsten-Edmands, V, Robinson, C.V, Rubinstein, J.L. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural comparison of the vacuolar and Golgi V-ATPases fromSaccharomyces cerevisiae.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6U7H
 
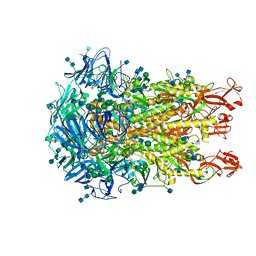 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
7TK0
 
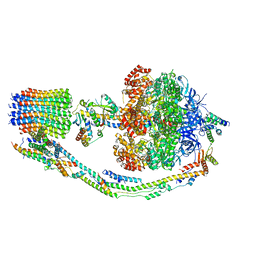 | |
7TJY
 
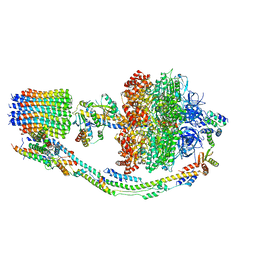 | |
7TJZ
 
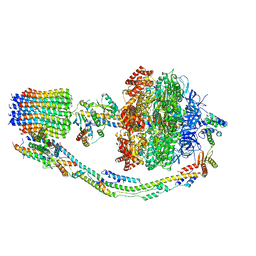 | |
7TK3
 
 | |
7TK2
 
 | |
7TK4
 
 | |
7TK1
 
 | |
