6Q6Q
 
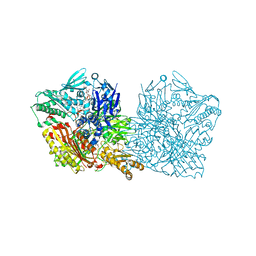 | | Human aldehyde oxidase SNP G1269R | | Descriptor: | Aldehyde oxidase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mota, C, Coelho, C, Santos-Silva, T, Romao, M.J. | | Deposit date: | 2018-12-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.10003781 Å) | | Cite: | Human aldehyde oxidase (hAOX1): structure determination of the Moco-free form of the natural variant G1269R and biophysical studies of single nucleotide polymorphisms.
Febs Open Bio, 9, 2019
|
|
3O5A
 
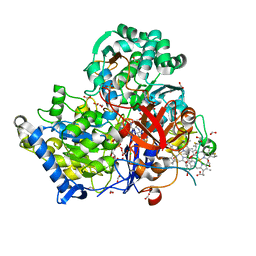 | | Crystal Structure of partially reduced Periplasmic Nitrate Reductase from Cupriavidus necator using Ionic Liquids | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
1VLB
 
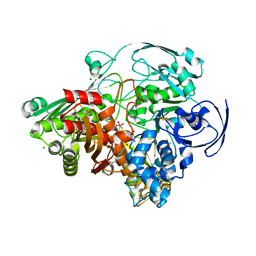 | | STRUCTURE REFINEMENT OF THE ALDEHYDE OXIDOREDUCTASE FROM DESULFOVIBRIO GIGAS AT 1.28 A | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Rebelo, J.M, Dias, J.M, Huber, R, Moura, J.J.G, Romao, M.J. | | Deposit date: | 2004-07-20 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structure refinement of the aldehyde oxidoreductase from Desulfovibrio gigas (MOP) at 1.28 A
J.Biol.Inorg.Chem., 6, 2001
|
|
8CM6
 
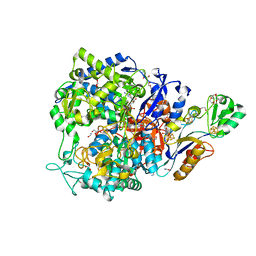 | | W-formate dehydrogenase C872A from Desulfovibrio vulgaris - with Formamide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, ... | | Authors: | Vilela-Alves, G, Mota, C, Oliveira, A.R, Manuel, R.R, Pereira, I.C, Romao, M.J. | | Deposit date: | 2023-02-17 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.416 Å) | | Cite: | An allosteric redox switch involved in oxygen protection in a CO 2 reductase.
Nat.Chem.Biol., 20, 2024
|
|
5NQD
 
 | | Arsenite oxidase AioAB from Rhizobium sp. str. NT-26 mutant AioBF108A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Santos-Silva, T, Romao, M, Vieira, M, Marques, A.T. | | Deposit date: | 2017-04-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arsenite oxidase AioAB from Rhizobium sp. str. NT-26 mutant AioBF108A
To Be Published
|
|
7OPN
 
 | | Human Aldehyde Oxidase SNP R1231H in complex with Raloxifene | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Aldehyde oxidase, DIMETHYL SULFOXIDE, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
7ORC
 
 | | Human Aldehyde Oxidase in complex with Raloxifene | | Descriptor: | Aldehyde oxidase, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Mota, C, Coelho, C, Santos Silva, T, Romao, M.J. | | Deposit date: | 2021-06-05 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interrogating the Inhibition Mechanisms of Human Aldehyde Oxidase by X-ray Crystallography and NMR Spectroscopy: The Raloxifene Case.
J.Med.Chem., 64, 2021
|
|
1SIJ
 
 | | Crystal structure of the Aldehyde Dehydrogenase (a.k.a. AOR or MOP) of Desulfovibrio gigas covalently bound to [AsO3]- | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ARSENITE, Aldehyde oxidoreductase, ... | | Authors: | Boer, D.R, Thapper, A, Brondino, C.D, Romao, M.J, Moura, J.J.G. | | Deposit date: | 2004-03-01 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Crystal Structure and EPR Spectra of "Arsenite-Inhibited" Desulfovibriogigas Aldehyde Dehydrogenase: A Member of the Xanthine Oxidase Family
J.Am.Chem.Soc., 126, 2004
|
|
1JAF
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C' FROM RHODOCYCLUS GELATINOSUS AT 2.5 ANGSTOMS RESOLUTION | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Archer, M, Banci, L, Dikaya, E, Romao, M.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cytochrome C' from Rhodocyclus Gelatinosus and Comparison with Other Cytochromes C'
J.Biol.Inorg.Chem., 2, 1997
|
|
5LU3
 
 | | The Structure of Spirochaeta thermophila CBM64 | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Correia, M.A.S, Romao, M.J, Carvalho, A.L. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C.
J. Biol. Chem., 292, 2017
|
|
6SDV
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Formate reduced form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, Formate dehydrogenase, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
6SDR
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Oxidized form | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, alpha subunit, ... | | Authors: | Oliveira, A.R, Mota, C, Mourato, C, Domingos, R.M, Santos, M.F.A, Gesto, D, Guigliarelli, B, Santos-Silva, T, Romao, M.J, Pereira, I.C. | | Deposit date: | 2019-07-29 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Towards the mechanistic understanding of enzymatic CO2 reduction
Acs Catalysis, 2020
|
|
5K39
 
 | | THE TYPE II COHESIN DOCKERIN COMPLEX FROM CLOSTRIDIUM THERMOCELLUM | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Dockerin module from a protein of unknown function | | Authors: | Viegas, A, Pinheiro, B, Bras, J.L.A, Romao, M.J, Alves, V, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-05-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
6QKN
 
 | | Structure of the azide-inhibited form of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae | | Descriptor: | AZIDE ION, CALCIUM ION, Cytochrome-c peroxidase, ... | | Authors: | Carvalho, A.L, Romao, M.J, Pauleta, S, Nobrega, C. | | Deposit date: | 2019-01-29 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mixed-valence, active form, of cytochrome c peroxidase from obligate human pathogenic bacterium Neisseria gonorrhoeae
To Be Published
|
|
4USA
 
 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with trans-cinnamaldehyde | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Aromatic Aldehydes at the Active Site of Aldehyde Oxidoreductase from Desulfovibrio Gigas: Reactivity and Molecular Details of the Enzyme-Substrate and Enzyme-Product Interaction.
J.Biol.Inorg.Chem., 20, 2015
|
|
2XJW
 
 | | Lysozyme-CO releasing molecule adduct | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos-Silva, T, Mukhopadhyay, A, Romao, M.J. | | Deposit date: | 2010-07-06 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Corm-3 Reactivity Toward Proteins: The Crystal Structure of a Ru(II) Dicarbonyl-Lysozyme Complex.
J.Am.Chem.Soc., 133, 2011
|
|
8RCB
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air (in a not degassed drop) for 34 min in the presence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement
Chem Sci, 2024
|
|
8RCC
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - aerobic soaked with 48 bar CO2 for 1 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBON DIOXIDE, ... | | Authors: | Vilela-Alves, G, Carpentier, P, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement
Chem Sci, 2024
|
|
8RC8
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 0 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement
Chem Sci, 2024
|
|
8RCA
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Reoxidized by exposure to air for 1 h in the absence of Formate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.664 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement
Chem Sci, 2024
|
|
8RC9
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - Co-crystallized with Formate and Exposed to air for 2 h | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Formate dehydrogenase, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement
Chem Sci, 2024
|
|
8RCG
 
 | | W-formate dehydrogenase M405S from Desulfovibrio vulgaris | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vilela-Alves, G, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Structural and biochemical characterization of the M405S variant of Desulfovibrio vulgaris formate dehydrogenase.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2W5F
 
 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2008-12-10 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
1Z1N
 
 | |
1RZ6
 
 | | Di-haem Cytochrome c Peroxidase, Form IN | | Descriptor: | CITRIC ACID, Cytochrome c peroxidase, HEME C | | Authors: | Dias, J.M, Alves, T, Bonifacio, C, Pereira, A, Bourgeois, D, Moura, I, Romao, M.J. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the mechanism of Ca(2+) activation of the di-heme cytochrome c peroxidase from Pseudomonas nautica 617.
Structure, 12, 2004
|
|
