2W2B
 
 | | Crystal Structure of single point mutant Tyr20Phe p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | ACETATE ION, ISOPROPYL ALCOHOL, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2W2F
 
 | | CRYSTAL STRUCTURE OF SINGLE POINT MUTANT ARG48GLN OF P-COUMARIC ACID DECARBOXYLASE FROM LACTOBACILLUS PLANTARUM STRUCTURAL INSIGHTS INTO THE ACTIVE SITE AND DECARBOXYLATION CATALYTIC MECHANISM | | Descriptor: | BARIUM ION, P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-29 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2W2A
 
 | | Crystal Structure of p-coumaric Acid Decarboxylase from Lactobacillus plantarum: structural insights into the active site and decarboxylation catalytic mechanism | | Descriptor: | P-COUMARIC ACID DECARBOXYLASE | | Authors: | Rodriguez, H, Angulo, I, de las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-10-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2WSJ
 
 | | Crystal Structure of single point mutant Glu71Ser p-coumaric Acid Decarboxylase | | Descriptor: | BARIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Rodriguez, H, Angulo, I, De Las Rivas, B, Campillo, N, Paez, J.A, Munoz, R, Mancheno, J.M. | | Deposit date: | 2009-09-07 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | P-Coumaric Acid Decarboxylase from Lactobacillus Plantarum: Structural Insights Into the Active Site and Decarboxylation Catalytic Mechanism.
Proteins, 78, 2010
|
|
2W37
 
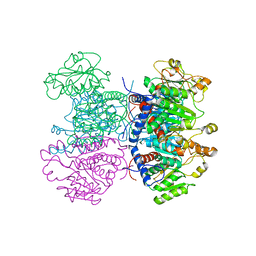 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
6YQ4
 
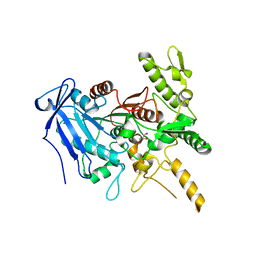 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
