5CZK
 
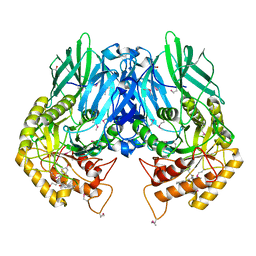 | | Structure of E. coli beta-glucuronidase bound with a novel, potent inhibitor 1-((6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea | | Descriptor: | 1-[(6,8-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(4-hydroxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Roberts, A.R, Wallace, B.R, Redinbo, M.R. | | Deposit date: | 2015-07-31 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure and Inhibition of Microbiome beta-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity.
Chem.Biol., 22, 2015
|
|
4HTG
 
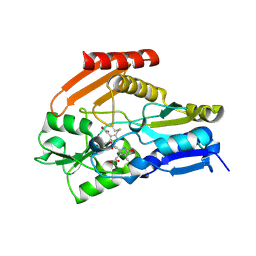 | | Porphobilinogen Deaminase from Arabidopsis Thaliana | | Descriptor: | 3-[(5Z)-5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methylidene}-4-(carboxymethyl)-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]propanoic acid, ACETATE ION, Porphobilinogen deaminase, ... | | Authors: | Roberts, A, Gill, R, Hussey, R.J, Erskine, P.T, Cooper, J.B, Wood, S.P, Chrystal, E.J.T, Shoolingin-Jordan, P.M. | | Deposit date: | 2012-11-01 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the mechanism of pyrrole polymerization catalysed by porphobilinogen deaminase: high-resolution X-ray studies of the Arabidopsis thaliana enzyme.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2G7H
 
 | |
4JHZ
 
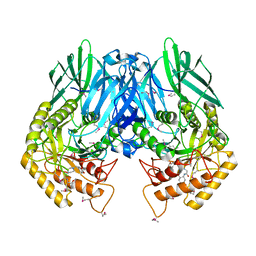 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide | | Descriptor: | 2-[4-(1,3-benzodioxol-5-ylmethyl)piperazin-1-yl]-N-[(1S,2S,5S)-2,5-dimethoxycyclohexyl]acetamide, Beta-glucuronidase | | Authors: | Roberts, A.B, Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.831 Å) | | Cite: | Molecular Insights into Microbial beta-Glucuronidase Inhibition to Abrogate CPT-11 Toxicity.
Mol.Pharmacol., 84, 2013
|
|
1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
5OK1
 
 | |
6QZG
 
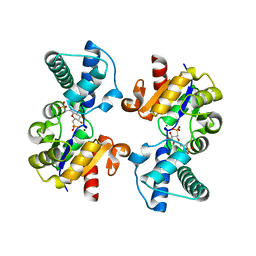 | |
6H8Y
 
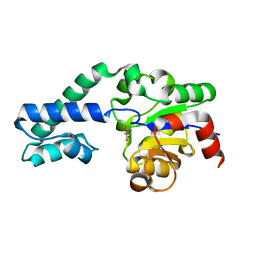 | |
6H91
 
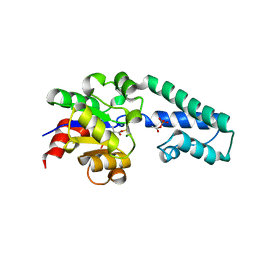 | |
6H90
 
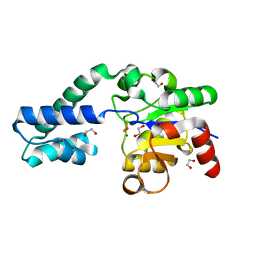 | |
6HDJ
 
 | |
6HDK
 
 | |
6HDM
 
 | | R49A variant of beta-phosphoglucomutase from Lactococcus lactis complexed with magnesium trifluoride and beta-G6P to 1.3 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-17 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6H8U
 
 | |
6H8W
 
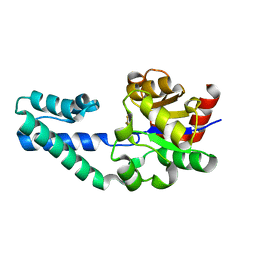 | |
6H92
 
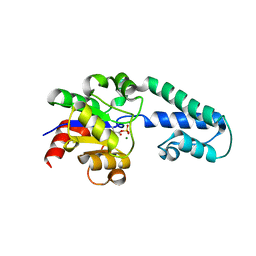 | |
6I03
 
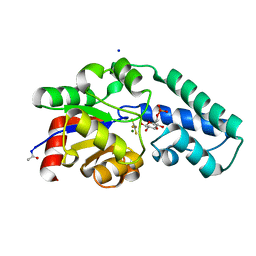 | |
6H8Z
 
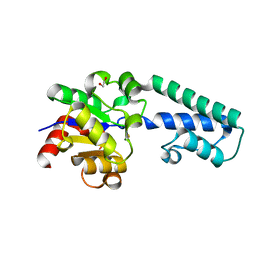 | |
5OK0
 
 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
5OK2
 
 | |
6H8V
 
 | | Beta-phosphoglucomutase from Lactococcus lactis in an open conformer in the P21 spacegroup to 1.8 A. | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6H94
 
 | | T16A variant of beta-phosphoglucomutase from Lactococcus lactis with phosphate and TRIS bound in an open conformer to 1.5 A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C, Waltho, J.P. | | Deposit date: | 2018-08-03 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Transition state of phospho-enzyme hydrolysis in beta-phosphoglucomutase.
To Be Published
|
|
6HDI
 
 | |
