4V8L
 
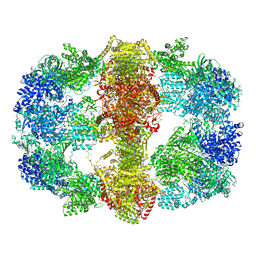 | |
2AGN
 
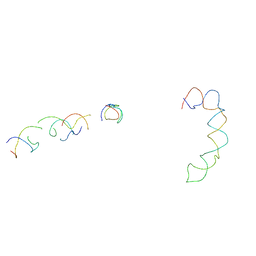 | | Fitting of hepatitis C virus internal ribosome entry site domains into the 15 A Cryo-EM map of a HCV IRES-80S ribosome (H. sapiens) complex | | Descriptor: | 6 nt A-RNA helix, HCV IRES DOMAIN II, HCV IRES IIIABC, ... | | Authors: | Boehringer, D, Thermann, R, Ostareck-Lederer, A, Lewis, J.D, Stark, H. | | Deposit date: | 2005-07-27 | | Release date: | 2006-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of the hepatitis C Virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES
Structure, 13, 2005
|
|
5K8Z
 
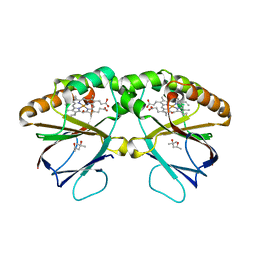 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 8.5) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Chlorite dismutase, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K90
 
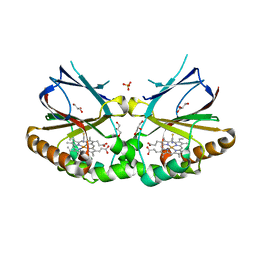 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with isothiocyanate | | Descriptor: | Chlorite dismutase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
5K91
 
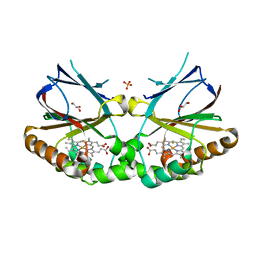 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 in complex with fluoride | | Descriptor: | Chlorite dismutase, FLUORIDE ION, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-31 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
4ADV
 
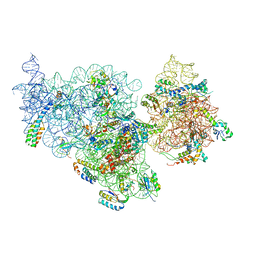 | | Structure of the E. coli methyltransferase KsgA bound to the E. coli 30S ribosomal subunit | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Boehringer, D, O'Farrell, H.C, Rife, J.P, Ban, N. | | Deposit date: | 2012-01-03 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structural Insights Into Methyltransferase Ksga Function in 30S Ribosomal Subunit Biogenesis
J.Biol.Chem., 287, 2012
|
|
9FEK
 
 | |
4Y19
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS18_alpha, FS18_beta, ... | | Authors: | Beringer, D.X, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
4BDX
 
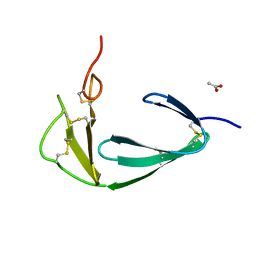 | |
8CLT
 
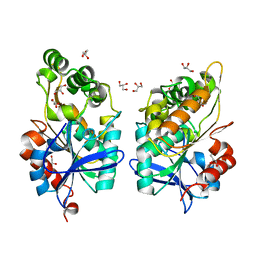 | |
8CLU
 
 | | Zearalenone lactonase from Rhodococcus erythropolis in complex with zearalactamenone | | Descriptor: | (4~{S})-4-methyl-16,18-bis(oxidanyl)-3-azabicyclo[12.4.0]octadeca-1(18),12,14,16-tetraene-2,8-dione, GLYCEROL, Zearalenone lactonase | | Authors: | Puehringer, D. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLO
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
5MAU
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 (pH 6.5) | | Descriptor: | Chlorite dismutase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
8CLP
 
 | |
5NKV
 
 | | Crystal structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 at pH 9.0 and 293 K. | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
4BDW
 
 | |
5NKU
 
 | | Joint neutron/X-ray structure of dimeric chlorite dismutase from Cyanothece sp. PCC7425 | | Descriptor: | CHLORIDE ION, Chlorite Dismutase, GLYCEROL, ... | | Authors: | Puehringer, D, Schaffner, I, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2017-04-03 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Molecular Mechanism of Enzymatic Chlorite Detoxification: Insights from Structural and Kinetic Studies.
ACS Catal, 7, 2017
|
|
8CLN
 
 | | Zearalenone lactonase from Streptomyces coelicoflavus, SeMet derivative for SAD phasing | | Descriptor: | Hydrolase | | Authors: | Puehringer, D, Grishkovskaya, I, Mlynek, G, Kostan, J. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial Lactonases ZenA with Noncanonical Structural Features Hydrolyze the Mycotoxin Zearalenone.
Acs Catalysis, 14, 2024
|
|
8CLQ
 
 | |
8CLV
 
 | |
4B41
 
 | | Crystal structure of an amyloid-beta binding single chain antibody G7 | | Descriptor: | ANTIBODY G7, CHLORIDE ION, GLYCEROL | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Crystal Structure of an Amyloid-Beta Binding Single Chain Antibody G7
To be Published
|
|
4B5E
 
 | | Crystal Structure of an amyloid-beta binding single chain antibody PS2-8 | | Descriptor: | GLYCEROL, PS2-8, SULFATE ION | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Amyloid-Beta Binding Single Chain Antibody Ps2-8
To be Published
|
|
5LOQ
 
 | | Structure of coproheme bound HemQ from Listeria monocytogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Puehringer, D, Mlynek, G, Hofbauer, S, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies.
FEBS J., 283, 2016
|
|
4Y1A
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS17_alpha, FS17_beta, ... | | Authors: | Beringer, D.X, Vivian, J.P, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
4N0B
 
 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
