3J41
 
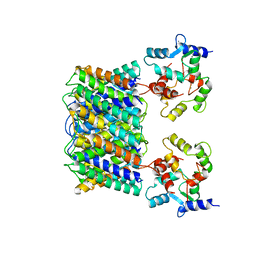 | | Pseudo-atomic model of the Aquaporin-0/Calmodulin complex derived from electron microscopy | | Descriptor: | CALCIUM ION, Calmodulin, Lens fiber major intrinsic protein | | Authors: | Reichow, S.L, Clemens, D.M, Freites, J.A, Nemeth-Cahalan, K.L, Heyden, M, Tobias, D.J, Hall, J.E, Gonen, T. | | Deposit date: | 2013-05-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Allosteric mechanism of water-channel gating by Ca(2+)-calmodulin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3J4Q
 
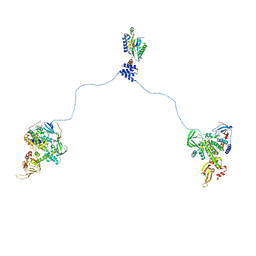 | |
3J4R
 
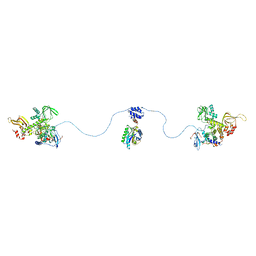 | |
5U6Y
 
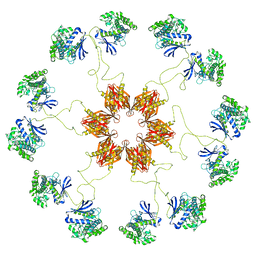 | | Pseudo-atomic model of the CaMKIIa holoenzyme. | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Myers, J, Reichow, S.L. | | Deposit date: | 2016-12-09 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | The CaMKII holoenzyme structure in activation-competent conformations.
Nat Commun, 8, 2017
|
|
6MHY
 
 | |
6MHQ
 
 | |
7JMD
 
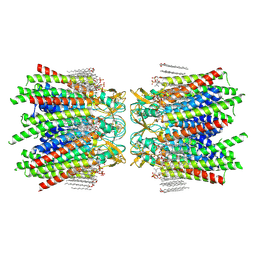 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JM9
 
 | | Sheep Connexin-50 at 2.5 angstroms reoslution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JLW
 
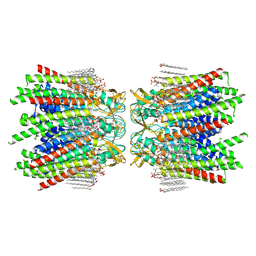 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-30 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN0
 
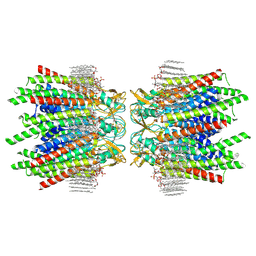 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 2 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.A, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JJP
 
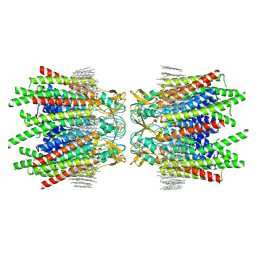 | | Sheep Connexin-50 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JN1
 
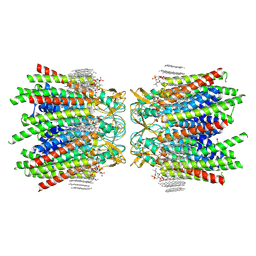 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-08-03 | | Release date: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JMC
 
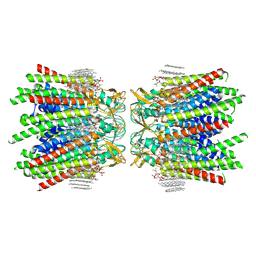 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2AQA
 
 | |
2AQC
 
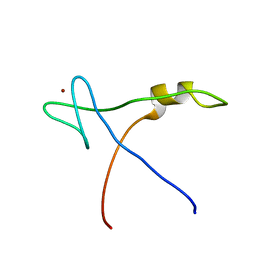 | | NMR Structural analysis of archaeal Nop10 | | Descriptor: | Ribosome biogenesis protein Nop10, ZINC ION | | Authors: | Hamma, T, Reichow, S.L, Varani, G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-08-17 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2APO
 
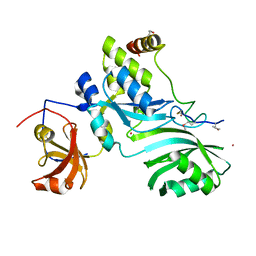 | | Crystal Structure of the Methanococcus jannaschii Cbf5 Nop10 Complex | | Descriptor: | POTASSIUM ION, Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, ... | | Authors: | Hamma, T, Reichow, S.L, Varani, G, Ferre-D'Amare, A.R. | | Deposit date: | 2005-08-16 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7JKC
 
 | | Sheep Connexin-46 at 1.9 angstroms resolution by CryoEM | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
