7LZI
 
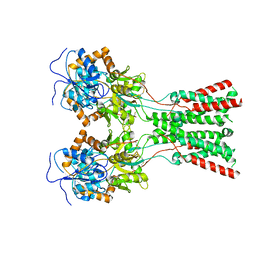 | |
7LZH
 
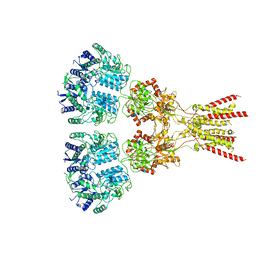 | | Structure of the glutamate receptor-like channel AtGLR3.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-09 | | Release date: | 2021-07-28 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ0
 
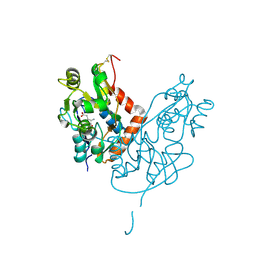 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with glutamate | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LZ1
 
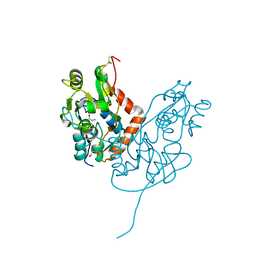 | | Structure of glutamate receptor-like channel GLR3.4 ligand-binding domain in complex with serine | | Descriptor: | GLYCEROL, Glutamate receptor 3.4, SERINE, ... | | Authors: | Gangwar, S.P, Green, M.N, Sobolevsky, A.I. | | Deposit date: | 2021-03-08 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure of the Arabidopsis thaliana glutamate receptor-like channel GLR3.4.
Mol.Cell, 81, 2021
|
|
7LTE
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTC
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTF
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant Y58F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTR
 
 | | Structure of the heteromeric complex between the alpha-N-methyltransferase (SonM) and a truncated construct of the RiPP precursor (SonA) (with SAM) | | Descriptor: | GLYCEROL, LigA domain-containing protein, S-ADENOSYLMETHIONINE, ... | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTH
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant Y93F) and RiPP precursor (SonA) heteromeric complex (no cofactor) | | Descriptor: | LigA domain-containing protein, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
7LTS
 
 | | Structure of the alpha-N-methyltransferase (SonM mutant R67A) and RiPP precursor (SonA) heteromeric complex (with SAH) | | Descriptor: | LigA domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, TP-methylase family protein | | Authors: | Miller, F.S, Crone, K.K, Jensen, M.R, Shaw, S, Harcombe, W.R, Elias, M, Freeman, M.F. | | Deposit date: | 2021-02-20 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Conformational rearrangements enable iterative backbone N-methylation in RiPP biosynthesis.
Nat Commun, 12, 2021
|
|
