2CJH
 
 | |
2CJG
 
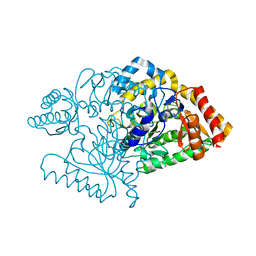 | | Lysine aminotransferase from M. tuberculosis in bound PMP form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-LYSINE-EPSILON AMINOTRANSFERASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2006-04-01 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Direct Evidence for a Glutamate Switch Necessary for Substrate Recognition: Crystal Structures of Lysine Epsilon-Aminotransferase (Rv3290C) from Mycobacterium Tuberculosis H37Rv.
J.Mol.Biol., 362, 2006
|
|
2CJD
 
 | |
3RB9
 
 | |
3S5C
 
 | | Crystal Structure of a Hexachlorocyclohexane dehydrochlorinase (LinA) Type2 | | Descriptor: | LinA | | Authors: | Kukshal, V, Macwan, A.S, Kumar, A, Ramachandran, R. | | Deposit date: | 2011-05-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the hexachlorocyclohexane dehydrochlorinase (LinA-type2): mutational analysis, thermostability and enantioselectivity
Plos One, 7, 2012
|
|
1SSN
 
 | | STAPHYLOKINASE, SAKSTAR VARIANT, NMR, 20 STRUCTURES | | Descriptor: | STAPHYLOKINASE | | Authors: | Ohlenschlager, O, Ramachandran, R, Guhrs, K.H, Schlott, B, Brown, L.R. | | Deposit date: | 1998-06-07 | | Release date: | 1998-12-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the plasminogen-activator protein staphylokinase.
Biochemistry, 37, 1998
|
|
1RFR
 
 | | NMR structure of the 30mer stemloop-D of coxsackieviral RNA | | Descriptor: | stemloop-D RNA of the 5'-cloverleaf of coxsackievirus B3 | | Authors: | Ohlenschlager, O, Wohnert, J, Bucci, E, Seitz, S, Hafner, S, Ramachandran, R, Zell, R, Gorlach, M. | | Deposit date: | 2003-11-10 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf
RNA and its interaction with the proteinase 3C.
STRUCTURE, 12, 2004
|
|
1ZAU
 
 | |
1UUU
 
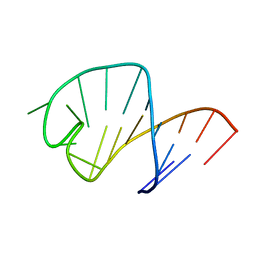 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1KMA
 
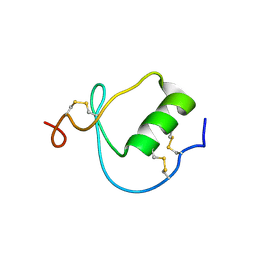 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
1S9L
 
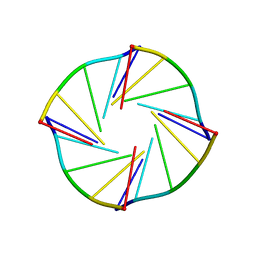 | | NMR Solution Structure of a Parallel LNA Quadruplex | | Descriptor: | 5'-((TLN)P*(LCG)P*(LCG)P*(LCG)P*(TLN))-3' | | Authors: | Randazzo, A, Esposito, V, Ohlenschlager, O, Ramachandran, R, Mayola, L. | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a parallel LNA quadruplex.
Nucleic Acids Res., 32, 2004
|
|
2W24
 
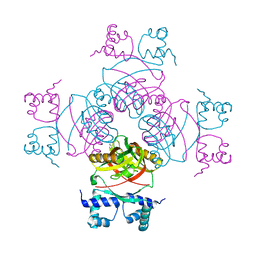 | | M. tuberculosis Rv3291c complexed to Lysine | | Descriptor: | LYSINE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Ramachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
2VOE
 
 | | Crystal structure of Rv2780 from M. tuberculosis H37Rv | | Descriptor: | ALANINE DEHYDROGENASE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-17 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins, 72, 2008
|
|
2W25
 
 | | Crystal structure of Glu104Ala mutant | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, RAmachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
2VU5
 
 | | Crystal structure of Pndk from Bacillus anthracis | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Misra, G, Aggarwal, A, Dube, D, Zaman, M.S, Singh, Y, Ramachandran, R. | | Deposit date: | 2008-05-21 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bacillus Anthracis Nucleoside Diphosphate Kinase and its Characterization Reveals an Enzyme Adapted to Perform Under Stress Conditions.
Proteins, 76, 2009
|
|
2VOJ
 
 | | Ternary complex of M. tuberculosis Rv2780 with NAD and pyruvate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, ALANINE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-02-18 | | Release date: | 2008-03-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Mycobacterium Tuberculosis Secretory Antigen Alanine Dehydrogenase (Rv2780) in Apo and Ternary Complex Forms Captures "Open" and "Closed" Enzyme Conformations.
Proteins: Struct., Funct., Bioinf., 72, 2008
|
|
1M4Y
 
 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
2VC0
 
 | |
2VBY
 
 | |
2VC1
 
 | |
2VBZ
 
 | |
2VBX
 
 | |
2VBW
 
 | |
2IVM
 
 | | Crystal structure of a transcriptional regulator | | Descriptor: | TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Ramachandran, R. | | Deposit date: | 2006-06-14 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Insights from the Crystal Structures of a Feast/Famine Regulatory Protein from Mycobacterium Tuberculosis H37Rv.
Nucleic Acids Res., 35, 2007
|
|
2JJF
 
 | | N328A mutant of M. tuberculosis Rv3290c | | Descriptor: | L-LYSINE EPSILON AMINOTRANSFERASE | | Authors: | tripathi, S.M, Ramachandran, R. | | Deposit date: | 2008-04-04 | | Release date: | 2009-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational Analysis of Mycobacterium Tuberculosis Lysine Epsilon-Aminotransferase and Inhibitor Co-Crystal Structures, Reveals Distinct Binding Modes.
Biochem.Biophys.Res.Commun., 463, 2015
|
|
