3IEK
 
 | | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-22 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of native TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
3IEL
 
 | | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R. | | Deposit date: | 2009-07-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of TTHA0252 from Thermus thermophilus HB8 complexed with UMP
To be Published
|
|
3IE1
 
 | | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA | | Descriptor: | CITRATE ANION, RNA (5'-R(P*UP*UP*UP*U)-3'), Ribonuclease TTHA0252, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of H380A mutant TTHA0252 from Thermus thermophilus HB8 complexed with RNA
To be Published
|
|
3IE2
 
 | | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of H400V mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
2ISM
 
 | | Crystal structure of the putative oxidoreductase (glucose dehydrogenase) (TTHA0570) from thermus theromophilus HB8 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative oxidoreductase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Ebihara, A, Shinkai, A, Nakagawa, N, Shimizu, N, Yamamoto, M, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-18 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Oxidoreductase (Glucose Dehydrogenase) (TTHA0570) from Thermus Theromophilus HB8
To be Published
|
|
1VIP
 
 | | ANTICOAGULANT CLASS II PHOSPHOLIPASE A2 FROM THE VENOM OF VIPERA RUSSELLI RUSSELLI | | Descriptor: | PHOSPHOLIPASE A2, SULFATE ION | | Authors: | Carredano, E, Westerlund, B, Persson, B, Saarinen, M, Ramaswamy, S, Eaker, D, Eklund, H. | | Deposit date: | 1997-02-27 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structures of two toxins from snake venom throw light on the anticoagulant and neurotoxic sites of phospholipase A2.
Toxicon, 36, 1998
|
|
2IIH
 
 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
2IRP
 
 | | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Putative aldolase class 2 protein aq_1979 | | Authors: | Jeyakanthan, J, Gayathri, D, Yogavel, M, Velmurugan, D, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5
To be Published
|
|
3IDZ
 
 | | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of S378Q mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
2IEX
 
 | | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426 | | Descriptor: | Dihydroxynapthoic acid synthetase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, BaBa, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426
To be Published
|
|
2IS8
 
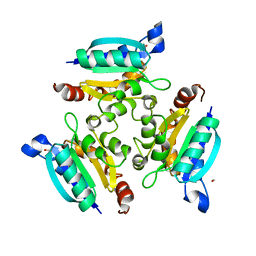 | | Crystal structure of the Molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | Descriptor: | FORMIC ACID, Molybdopterin biosynthesis enzyme, MoaB | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8
To be Published
|
|
1CQ6
 
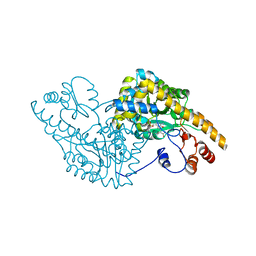 | | ASPARTATE AMINOTRANSFERASE COMPLEX WITH C4-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1I1X
 
 | |
1CQ7
 
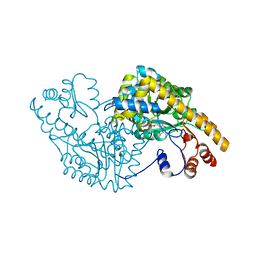 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C5-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
2IDE
 
 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
1JU9
 
 | |
3IE0
 
 | | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of S378Y mutant TTHA0252 from Thermus thermophilus HB8
To be Published
|
|
1GC3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH TRYPTOPHAN | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
2III
 
 | | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5
To be Published
|
|
1GCK
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-08-04 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GD7
 
 | | CRYSTAL STRUCTURE OF A BIFUNCTIONAL PROTEIN (CSAA) WITH EXPORT-RELATED CHAPERONE AND TRNA-BINDING ACTIVITIES. | | Descriptor: | CSAA PROTEIN | | Authors: | Shibata, T, Inoue, Y, Vassylyev, D.G, Kawaguchi, S, Yokoyama, S, Muller, J, Linde, D, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.
EMBO J., 20, 2001
|
|
1XSM
 
 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
1GC4
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1YX9
 
 | |
1ZB1
 
 | | Structure basis for endosomal targeting by the Bro1 domain | | Descriptor: | BRO1 protein | | Authors: | Kim, J, Sitaraman, S, Hierro, A, Beach, B.M, Odorizzi, G, Hurley, J.H. | | Deposit date: | 2005-04-07 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for endosomal targeting by the Bro1 domain.
Dev.Cell, 8, 2005
|
|
