8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
7V0J
 
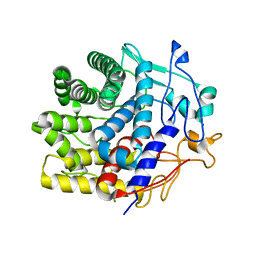 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellobiose product | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7UNP
 
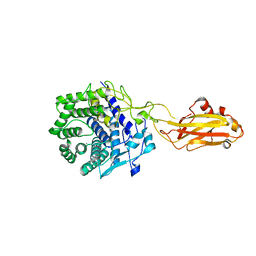 | | Crystal structure of the CelR catalytic domain and CBM3c | | Descriptor: | CALCIUM ION, Glucanase | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7V0I
 
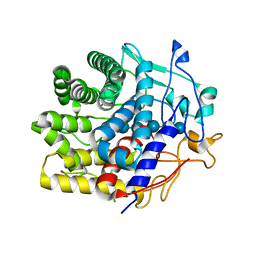 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
8AHW
 
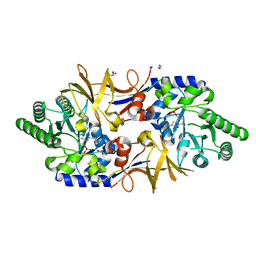 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8PZQ
 
 | | Model for focused reconstruction of influenza A RNP-like particle | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
8PZP
 
 | | Model for influenza A virus helical ribonucleoprotein-like structure | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
8B8H
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from M. tuberculosis in complex with DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, Alanine racemase, ... | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
8B61
 
 | | Crystal structure of BfrC protein from Bacteroides fragilis NCTC 9343 | | Descriptor: | Conserved hypothetical lipoprotein, GLYCEROL, pentane-1,3,5-tricarboxylic acid | | Authors: | Antonyuk, S.V, Barnett, K, Strange, R.W, Olczak, T. | | Deposit date: | 2022-09-25 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Bacteroides fragilis expresses three proteins similar to Porphyromonas gingivalis HmuY: Hemophore-like proteins differentially evolved to participate in heme acquisition in oral and gut microbiomes.
Faseb J., 37, 2023
|
|
8B6A
 
 | |
8PWS
 
 | | Dye-decolourising peroxidase DtpB (56 kGy) | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Owen, R.L, Strange, R.W. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dye-decolourising peroxidase DtpB (56 kGy)
To Be Published
|
|
8PWY
 
 | | Dye-decolourising peroxidase DtpB (112 kGy) | | Descriptor: | MAGNESIUM ION, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lucic, M, Worrall, J.A.R, Hough, M.A, Owen, R.L, Strange, R.W. | | Deposit date: | 2023-07-21 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dye-decolourising peroxidase DtpB (56 kGy)
To Be Published
|
|
3ND1
 
 | |
3NEI
 
 | |
6BF2
 
 | |
3NDC
 
 | |
6UI3
 
 | | GH5-4 broad specificity endoglucanase from Clostridum cellulovorans | | Descriptor: | 1,2-ETHANEDIOL, Cellulase | | Authors: | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
3NDY
 
 | |
3O6B
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
4WFW
 
 | |
3NUT
 
 | |
4X6E
 
 | | CD1a binary complex with lysophosphatidylcholine | | Descriptor: | (4R,7R,18Z)-4,7-dihydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphaheptacos-18-en-1-aminium 4-oxide, Beta-2-microglobulin, D-MALATE, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-12-08 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | alpha beta T cell antigen receptor recognition of CD1a presenting self lipid ligands.
Nat.Immunol., 16, 2015
|
|
3O2U
 
 | | S. cerevisiae Ubc12 | | Descriptor: | GLYCEROL, NEDD8-conjugating enzyme UBC12 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
6CNH
 
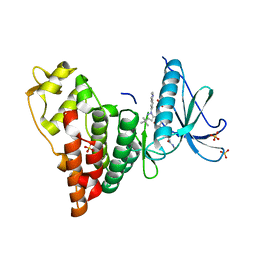 | | Human PRPF4B in complex with Rebastinib | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Godoi, P.H.C, Santiago, A.S, Ramos, P.Z, Fala, A.M, Salmazo, A.P.T, Counago, R.M, Righetto, G.L, Silva, P.N.B, Gileadi, O, Guimaraes, C.R.W, Massirer, K.B, Arruda, P, Elkins, J.M, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human PRPF4B in complex with Rebastinib
To be Published
|
|
1AAB
 
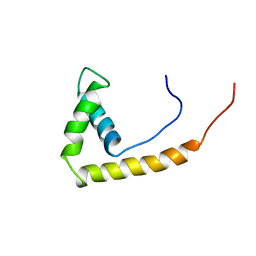 | | NMR STRUCTURE OF RAT HMG1 HMGA FRAGMENT | | Descriptor: | HIGH MOBILITY GROUP PROTEIN | | Authors: | Hardman, C.H, Broadhurst, R.W, Raine, A.R.C, Grasser, K.D, Thomas, J.O, Laue, E.D. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 34, 1995
|
|
