5V0Z
 
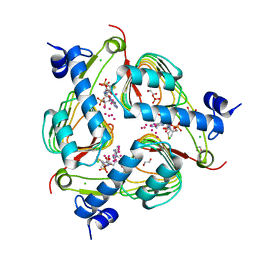 | | Crystal structure of Galactoside O-acetyltransferase complex with CoA (P32 space group). | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Knapik, A.A, Niedzialkowska, E, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of Galactoside O-acetyltransferase complex with CoA (P32 space group).
to be published
|
|
3T7B
 
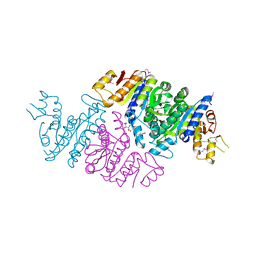 | | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis | | Descriptor: | Acetylglutamate kinase, GLUTAMIC ACID, S,R MESO-TARTARIC ACID | | Authors: | Demas, M.W, Solberg, R.G, Cooper, D.R, Chruszcz, M, Porebski, P.J, Zheng, H, Onopriyenko, O, Skarina, T, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-29 | | Release date: | 2011-09-14 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of N-acetyl-L-glutamate kinase from Yersinia pestis
To be Published
|
|
3V03
 
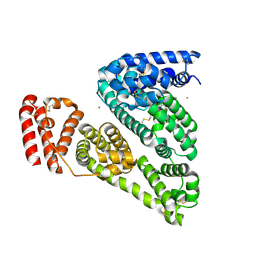 | | Crystal structure of Bovine Serum Albumin | | Descriptor: | ACETATE ION, CALCIUM ION, Serum albumin | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3UEE
 
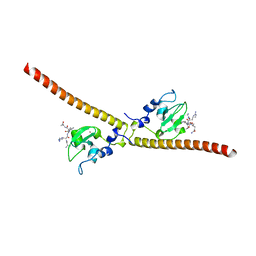 | | Crystal structure of human Survivin K62A mutant bound to N-terminal histone H3 | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UED
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3 (C2 space group). | | Descriptor: | Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEH
 
 | | Crystal structure of human Survivin H80A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3V09
 
 | | Crystal structure of Rabbit Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3V08
 
 | | Crystal structure of Equine Serum Albumin | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, SULFATE ION, ... | | Authors: | Dayal, A, Jablonska, K, Porebski, P.J, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-07 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and immunologic characterization of bovine, horse, and rabbit serum albumins.
Mol.Immunol., 52, 2012
|
|
3UEG
 
 | | Crystal structure of human Survivin K62A mutant | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, TETRAETHYLENE GLYCOL, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEC
 
 | | Crystal structure of human Survivin bound to histone H3 phosphorylated on threonine-3. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Baculoviral IAP repeat-containing protein 5, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Cooper, D.R, Chruszcz, M, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3UEI
 
 | | Crystal structure of human Survivin E65A mutant | | Descriptor: | Baculoviral IAP repeat-containing protein 5, ZINC ION | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol Biol Cell, 23, 2012
|
|
3UEF
 
 | | Crystal structure of human Survivin bound to histone H3 (C2 space group). | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 5, N-terminal fragment of histone H3, ... | | Authors: | Niedzialkowska, E, Porebski, P.J, Wang, F, Higgins, J.M, Stukenberg, P.T, Minor, W. | | Deposit date: | 2011-10-30 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres.
Mol.Biol.Cell, 23, 2012
|
|
3TPF
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4Z0P
 
 | | Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase SMc02828 (SmGhrA) from Sinorhizobium meliloti in complex with NADPH and oxalate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sroka, P, Gasiorowska, O.A, Handing, K.B, Shabalin, I.G, Porebski, P.J, Hillerich, B.S, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-26 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry, 57, 2018
|
|
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KPS
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
4KOW
 
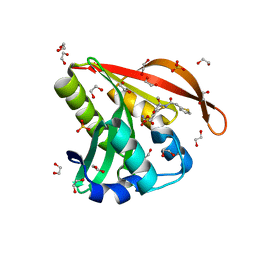 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefoxitin | | Descriptor: | 1,2-ETHANEDIOL, CEFOXITIN, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Grabowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOY
 
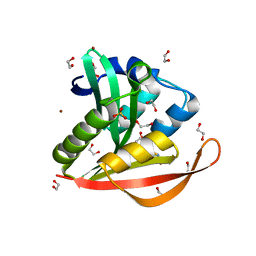 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KLW
 
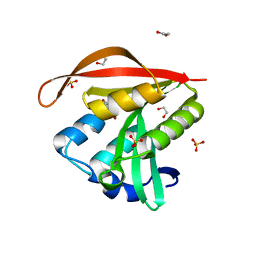 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 2-(aminocarbonyl)benzoate | | Descriptor: | 1,2-ETHANEDIOL, 2-carbamoylbenzoic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4KOR
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with 7-aminocephalosporanic acid | | Descriptor: | 1,2-ETHANEDIOL, 7-aminocephalosporanic acid, SULFATE ION, ... | | Authors: | Majorek, K.A, Porebski, P.J, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
3V48
 
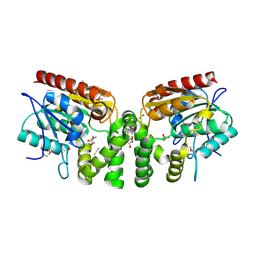 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
3V4D
 
 | | Crystal structure of RutC protein a member of the YjgF family from E.coli | | Descriptor: | Aminoacrylate peracid reductase RutC | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Escherichia coli RutC, a member of the YjgF family and putative aminoacrylate peracid reductase of the rut operon.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
