8ZBO
 
 | | Crystal structure of the biphotochromic fluorescent protein moxSAASoti (F97M variant) in its green on-state | | Descriptor: | 1,2-ETHANEDIOL, F97M variant of the biphotochromic fluorescent protein moxSAASoti, NITRATE ION, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Marynich, N.K, Minyaev, M.E, Khadiyatova, A.A, Popov, V.O, Savitsky, A.P. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single-point substitution F97M leads to in cellulo crystallization of the biphotochromic protein moxSAASoti.
Biochem.Biophys.Res.Commun., 732, 2024
|
|
7P8O
 
 | | Crystal structure of D-aminoacid transaminase from Haliscomenobacter hydrossis in its intermediate form | | Descriptor: | Aminotransferase class IV, MAGNESIUM ION, SULFATE ION | | Authors: | Matyuta, I.O, Boyko, K.M, Bakunova, A.K, Nikolaeva, A.Y, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Incorporation of pyridoxal-5'-phosphate into the apoenzyme: A structural study of D-amino acid transaminase from Haliscomenobacter hydrossis.
Biochim Biophys Acta Proteins Proteom, 1873, 2024
|
|
5CE8
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis | | Descriptor: | Branched-chain amino acid aminotransferase, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-06 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | First structure of archaeal branched-chain amino acid aminotransferase from Thermoproteus uzoniensis specific for L-amino acids and R-amines.
Extremophiles, 20, 2016
|
|
7PPP
 
 | | Crystal structure of ZAD-domain of ZNF_276 protein from rabbit. | | Descriptor: | ZINC ION, Zinc finger protein 276 | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-14 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POH
 
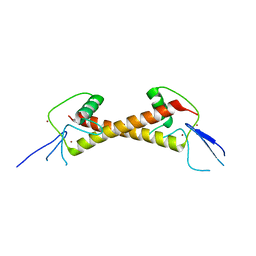 | | Crystal structure of ZAD-domain of Serendipity-d protein from D.melanogaster | | Descriptor: | Serendipity locus protein delta, ZINC ION | | Authors: | Boyko, K.M, Kachalova, G.S, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7POK
 
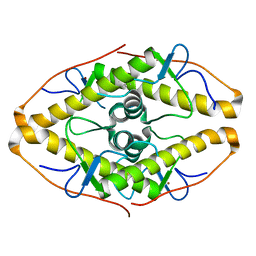 | | Crystal structure of ZAD-domain of Pita protein from D.melanogaster | | Descriptor: | LD15650p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
7PO9
 
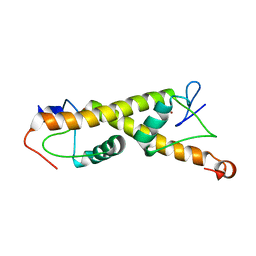 | | Crystal structure of ZAD-domain of M1BP protein from D.melanogaster | | Descriptor: | LD30467p, ZINC ION | | Authors: | Boyko, K.M, Bonchuk, A.N, Nikolaeva, A.Y, Georgiev, P.G, Popov, V.O. | | Deposit date: | 2021-09-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into highly similar spatial organization of zinc-finger associated domains with a very low sequence similarity.
Structure, 30, 2022
|
|
5CM0
 
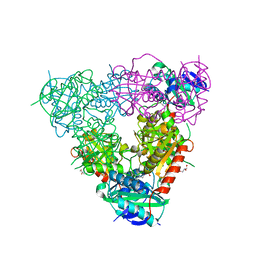 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans | | Descriptor: | Branched-chain transaminase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-07-16 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
7QGK
 
 | | The mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer in its red state | | Descriptor: | MAGNESIUM ION, The red form of the mRubyFT protein, Genetically Encoded Blue-to-Red Fluorescent Timer | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Gaivoronskii, F.A, Vlaskina, A.V, Subach, O.M, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-12-08 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer.
Int J Mol Sci, 23, 2022
|
|
7OIN
 
 | | Crystal structure of LSSmScarlet - a genetically encoded red fluorescent protein with a large Stokes shift | | Descriptor: | LSSmScarlet - Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift, SODIUM ION, SULFATE ION | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Dorovatovskii, P.V, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Ivashkina, O.I, Popov, V.O, Subach, F.V. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift.
Int J Mol Sci, 22, 2021
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5E25
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Geoglobus acetivorans complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, PYRIDOXAL-5'-PHOSPHATE, branched-chain aminotransferase | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Stekhanova, T.N, Mardanov, A.V, Rakitin, A.L, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermostable Branched-Chain Amino Acid Transaminases From the Archaea Geoglobus acetivorans and Archaeoglobus fulgidus : Biochemical and Structural Characterization.
Front Bioeng Biotechnol, 7, 2019
|
|
5EKC
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Shabalin, I.G, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure of thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+
To Be Published
|
|
5EK6
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 complexed with NADP and isobutyraldehyde | | Descriptor: | 2-methylpropanal, Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Polyakov, K.M, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5F2C
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 crystallized in microgravity (complex with NADP+) | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-12-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
5EUY
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
3SXQ
 
 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio paradoxus | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Boyko, K.M, Popov, V.O. | | Deposit date: | 2011-07-15 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
3SQR
 
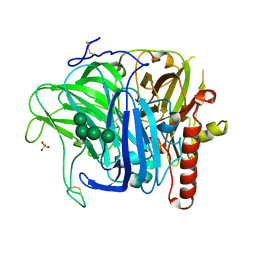 | | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, SULFATE ION, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-07-06 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3V9E
 
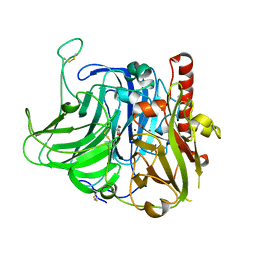 | | Structure of the L499M mutant of the laccase from B.aclada | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-12-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TTB
 
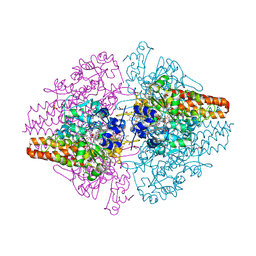 | | Structure of the Thioalkalivibrio paradoxus cytochrome c nitrite reductase in complex with sulfite | | Descriptor: | CALCIUM ION, COBALT (II) ION, Eight-heme nitrite reductase, ... | | Authors: | Polyakov, K.M, Trofimov, A.A, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-09-14 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural and functional analysis of two octaheme nitrite reductases from closely related Thioalkalivibrio species.
Febs J., 279, 2012
|
|
4FFK
 
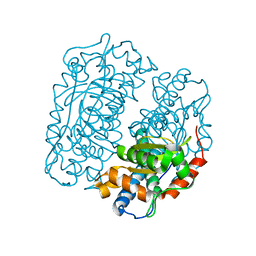 | | X-ray structure of iron superoxide dismutase from Acidilobus saccharovorans | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Safonova, T.N, Slutskaya, E.S, Dorovatovsky, P.V, Bezsudnova, E.Yu, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2012-06-01 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-ray structure of iron superoxide dismutase from Acidilobus saccharovorans
TO BE PUBLISHED
|
|
7Z79
 
 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | Descriptor: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
8AHU
 
 | | Crystal structure of D-amino acid aminotrensferase from Haliscomenobacter hydrossis complexed with D-cycloserine | | Descriptor: | Aminotransferase class IV, GLYCEROL, [5-hydroxy-6-methyl-4-({[(4E)-3-oxo-1,2-oxazolidin-4-ylidene]amino}methyl)pyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-22 | | Release date: | 2022-08-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Mechanism of D-Cycloserine Inhibition of D-Amino Acid Transaminase from Haliscomenobacter hydrossis.
Biochemistry Mosc., 88, 2023
|
|
7ZVR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
