6X8N
 
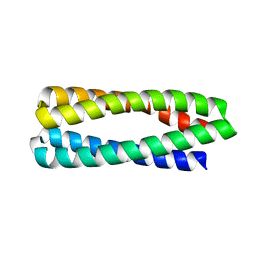 | | Crystal Structure of H49A ABLE mutant | | 分子名称: | De novo designed ABLE protein | | 著者 | Polizzi, N.F. | | 登録日 | 2020-06-01 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6W6X
 
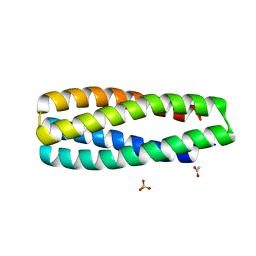 | | Crystal Structure of ABLE Apo-protein | | 分子名称: | ACETATE ION, De novo designed ABLE protein, SULFATE ION | | 著者 | Polizzi, N.F. | | 登録日 | 2020-03-18 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.297 Å) | | 主引用文献 | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
6W70
 
 | | Crystal Structure of apixaban-bound ABLE | | 分子名称: | 1-(4-METHOXYPHENYL)-7-OXO-6-[4-(2-OXOPIPERIDIN-1-YL)PHENYL]-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDINE-3-CARBOXAMIDE, ACETATE ION, De novo designed ABLE, ... | | 著者 | Polizzi, N.F. | | 登録日 | 2020-03-18 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.296 Å) | | 主引用文献 | A defined structural unit enables de novo design of small-molecule-binding proteins.
Science, 369, 2020
|
|
5TGY
 
 | | NMR structure of holo-PS1 | | 分子名称: | PS1, [5,10,15,20-tetrakis(trifluoromethyl)porphyrinato(2-)-kappa~4~N~21~,N~22~,N~23~,N~24~]zinc | | 著者 | Polizzi, N.F, Wu, Y. | | 登録日 | 2016-09-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
5TGW
 
 | | NMR structure of apo-PS1 | | 分子名称: | PS1 | | 著者 | Polizzi, N.F, Wu, Y. | | 登録日 | 2016-09-28 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a hyperstable non-natural protein-ligand complex with sub- angstrom accuracy.
Nat Chem, 9, 2017
|
|
