6H8N
 
 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis - mutant D285S | | Descriptor: | GLYCEROL, PHOSPHATE ION, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ... | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
6H8L
 
 | | Structure of peptidoglycan deacetylase PdaC from Bacillus subtilis | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan-N-acetylmuramic acid deacetylase PdaC, ZINC ION | | Authors: | Sainz-Polo, M.A, Grifoll-Romero, L, Albesa-Jove, D, Planas, A, Guerin, M.E. | | Deposit date: | 2018-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure-function relationships underlying the dualN-acetylmuramic andN-acetylglucosamine specificities of the bacterial peptidoglycan deacetylase PdaC.
J.Biol.Chem., 294, 2019
|
|
3D6E
 
 | | Crystal structure of the engineered 1,3-1,4-beta-glucanase protein from Bacillus licheniformis | | Descriptor: | Beta-glucanase, CALCIUM ION | | Authors: | Fita, I, Planas, A, Calisto, B.M, Addington, T. | | Deposit date: | 2008-05-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Re-engineering specificity in 1,3-1,4-beta-glucanase to accept branched xyloglucan substrates
Proteins, 79, 2011
|
|
2BVD
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | (3R,4R,5R)-4-hydroxy-5-(hydroxymethyl)piperidin-3-yl beta-D-glucopyranoside, ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-27 | | Release date: | 2005-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
2BV9
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-23 | | Release date: | 2005-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
6YUG
 
 | | Crystal structure of C. parvum GNA1 in complex with acetyl-CoA and glucose 6P. | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ACETYL COENZYME *A, Diamine acetyltransferase | | Authors: | Chi, J, Cova, M, de las Rivas, M, Medina, A, Borges, R, Leivar, P, Planas, A, Uson, I, Hurtado-Guerrero, R, Izquierdo, L. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Plasmodium falciparum Apicomplexan-Specific Glucosamine-6-Phosphate N -Acetyltransferase Is Key for Amino Sugar Metabolism and Asexual Blood Stage Development.
Mbio, 11, 2020
|
|
1U0A
 
 | | Crystal structure of the engineered beta-1,3-1,4-endoglucanase H(A16-M) in complex with beta-glucan tetrasaccharide | | Descriptor: | Beta-glucanase, CALCIUM ION, ZINC ION, ... | | Authors: | Gaiser, O.J, Piotukh, K, Ponnuswamy, M.N, Planas, A, Borriss, R, Heinemann, U. | | Deposit date: | 2004-07-13 | | Release date: | 2005-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis for the Substrate Specificity of a Bacillus 1,3-1,4-beta-Glucanase
J.Mol.Biol., 357, 2006
|
|
4NZ1
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ3
 
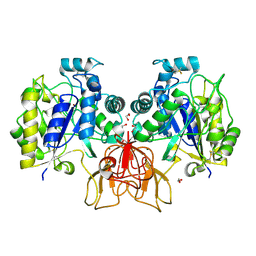 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ5
 
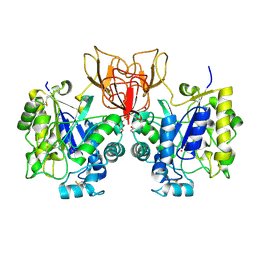 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with 2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE (NDG) and cadmium ion | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ACETATE ION, CADMIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Vibrio cholerae protein
To be Published
|
|
4OUI
 
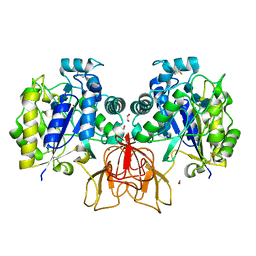 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with TRIACETYLCHITOTRIOSE (CTO) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NYY
 
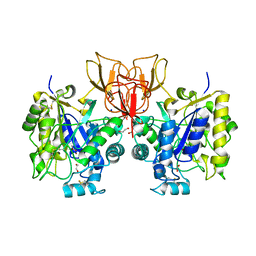 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NY2
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ4
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with 2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE (NDG) and zinc ion | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NYU
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in C 2 2 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
