3OD2
 
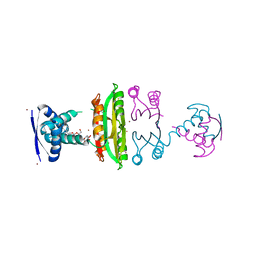 | | E. coli NikR soaked with excess nickel ions | | Descriptor: | 3-CYCLOHEXYLPROPYL 4-O-ALPHA-D-GLUCOPYRANOSYL-BETA-D-GLUCOPYRANOSIDE, NICKEL (II) ION, Nickel-responsive regulatory protein | | Authors: | Phillips, C.M, Schreiter, E.R, Drennan, C.L. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of low-affinity nickel binding to the nickel-responsive transcription factor NikR from Escherichia coli.
Biochemistry, 49, 2010
|
|
3BKT
 
 | |
3BKF
 
 | |
3BKU
 
 | |
4EIS
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, PEROXIDE ION, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
4EIR
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
