2R63
 
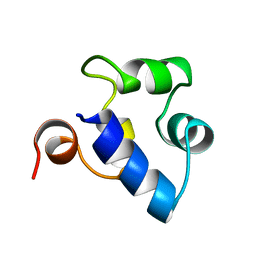 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-13 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1BHA
 
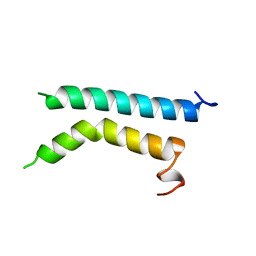 | | THREE-DIMENSIONAL STRUCTURE OF (1-71) BACTERIOOPSIN SOLUBILIZED IN METHANOL-CHLOROFORM AND SDS MICELLES DETERMINED BY 15N-1H HETERONUCLEAR NMR SPECTROSCOPY | | 分子名称: | BACTERIORHODOPSIN | | 著者 | Pervushin, K.V, Orekhov, V.Y, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | 登録日 | 1993-10-11 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | 著者 | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | 登録日 | 1996-11-08 | | 公開日 | 1997-06-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
4IMO
 
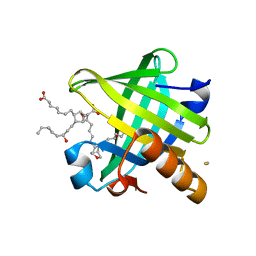 | | Crystal structure of wild type human Lipocalin PGDS in complex with substrate analog U44069 | | 分子名称: | (5E)-7-{(1R,4S,5S,6R)-5-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-6-yl}hept-5-enoic acid, Lipocalin-type prostaglandin-D synthase, THIOCYANATE ION | | 著者 | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | 登録日 | 2013-01-03 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
4IMN
 
 | | Crystal structure of wild type human Lipocalin PGDS bound with PEG MME 2000 | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Lipocalin-type prostaglandin-D synthase | | 著者 | Lim, S.M, Chen, D, Teo, H, Roos, A, Nyman, T, Tresaugues, L, Pervushin, K, Nordlund, P. | | 登録日 | 2013-01-03 | | 公開日 | 2013-03-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Structural and dynamic insights into substrate binding and catalysis of human lipocalin prostaglandin D synthase.
J.Lipid Res., 54, 2013
|
|
8HGA
 
 | |
8HIA
 
 | |
1BHB
 
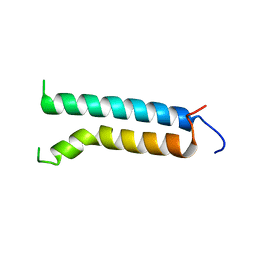 | | Three-dimensional structure of (1-71) bacterioopsin solubilized in methanol-chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy | | 分子名称: | BACTERIORHODOPSIN | | 著者 | Orekhov, V.Y, Pervushin, K.V, Popov, A.I, Musina, L.Y, Arseniev, A.S. | | 登録日 | 1993-10-11 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional structure of (1-71)bacterioopsin solubilized in methanol/chloroform and SDS micelles determined by 15N-1H heteronuclear NMR spectroscopy.
Eur.J.Biochem., 219, 1994
|
|
5WY9
 
 | |
1SR3
 
 | |
2JMM
 
 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | 分子名称: | Outer membrane protein A | | 著者 | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | 登録日 | 2006-11-20 | | 公開日 | 2007-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
2L54
 
 | | Solution structure of the Zalpha domain mutant of ADAR1 (N43A,Y47A) | | 分子名称: | Double-stranded RNA-specific adenosine deaminase | | 著者 | Zhao, J, Pervushin, K, Feng, S, Droge, P. | | 登録日 | 2010-10-24 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Alternate rRNA secondary structures as regulators of translation
Nat.Struct.Mol.Biol., 18, 2011
|
|
2K6I
 
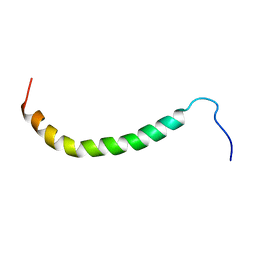 | | The domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H1-47 | | 分子名称: | Uncharacterized protein MJ0223 | | 著者 | Biukovic, N, Gayen, S, Pervushin, K, Gruber, G, Biukovic, G. | | 登録日 | 2008-07-09 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biophys.J., 97, 2009
|
|
2M1C
 
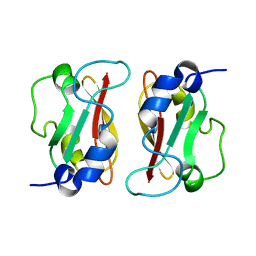 | | HADDOCK structure of GtYybT PAS Homodimer | | 分子名称: | DHH subfamily 1 protein | | 著者 | Liang, Z.X, Pervushin, K, Tan, E, Rao, F, Pasunooti, S, Soehano, I, Lescar, J. | | 登録日 | 2012-11-25 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the PAS Domain of a Thermophilic YybT Protein Homolog Reveals a Potential Ligand-binding Site.
J.Biol.Chem., 288, 2013
|
|
2LGT
 
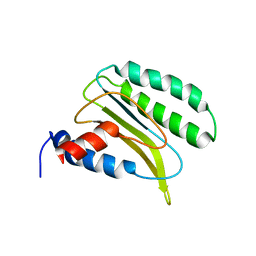 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | 分子名称: | Eukaryotic peptide chain release factor subunit 1 | | 著者 | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | 登録日 | 2011-08-02 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
2MQ6
 
 | |
2MI9
 
 | |
2MQ9
 
 | |
2MIA
 
 | |
