1C9O
 
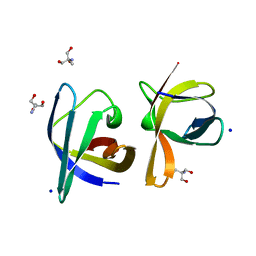 | | CRYSTAL STRUCTURE ANALYSIS OF THE BACILLUS CALDOLYTICUS COLD SHOCK PROTEIN BC-CSP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COLD-SHOCK PROTEIN, SODIUM ION | | Authors: | Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Thermal stability and atomic-resolution crystal structure of the Bacillus caldolyticus cold shock protein.
J.Mol.Biol., 297, 2000
|
|
1I5F
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD-SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-02-27 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZ9
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZA
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZB
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1HZC
 
 | | BACILLUS CALDOLYTICUS COLD-SHOCK PROTEIN MUTANTS TO STUDY DETERMINANTS OF PROTEIN STABILITY | | Descriptor: | COLD SHOCK PROTEIN CSPB, SODIUM ION | | Authors: | Delbrueck, H, Mueller, U, Perl, D, Schmid, F.X, Heinemann, U. | | Deposit date: | 2001-01-24 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal structures of mutant forms of the Bacillus caldolyticus cold shock protein differing in thermal stability.
J.Mol.Biol., 313, 2001
|
|
1KKS
 
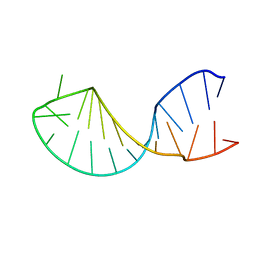 | | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression | | Descriptor: | 5'-R(*GP*GP*AP*AP*GP*GP*CP*CP*CP*UP*UP*UP*UP*CP*AP*GP*GP*GP*CP*CP*AP*CP*CP*C)-3' | | Authors: | Zanier, K, Luyten, I, Crombie, C, Muller, B, Schuemperli, D, Linge, J.P, Nilges, M, Sattler, M. | | Deposit date: | 2001-12-10 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression.
RNA, 8, 2002
|
|
1SMQ
 
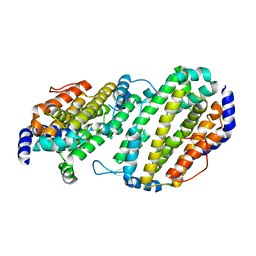 | | Structure of the Ribonucleotide Reductase Rnr2 Homodimer from Saccharomyces cerevisiae | | Descriptor: | Ribonucleoside-diphosphate reductase small chain 1 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
1SMS
 
 | | Structure of the Ribonucleotide Reductase Rnr4 Homodimer from Saccharomyces cerevisiae | | Descriptor: | MERCURY (II) ION, Ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Sommerhalter, M, Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers.
Biochemistry, 43, 2004
|
|
1JK0
 
 | | Ribonucleotide reductase Y2Y4 heterodimer | | Descriptor: | ZINC ION, ribonucleoside-diphosphate reductase small chain 1, ribonucleoside-diphosphate reductase small chain 2 | | Authors: | Voegtli, W.C, Perlstein, D.L, Ge, J, Stubbe, J, Rosenzweig, A.C. | | Deposit date: | 2001-07-10 | | Release date: | 2001-09-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the yeast ribonucleotide reductase Y2Y4 heterodimer.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
