2BIG
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure I) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI3
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure D) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI2
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure C) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIE
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure H) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-21 | | Release date: | 2005-05-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BIA
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure G) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI9
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure F) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI1
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure B) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BI5
 
 | | Radiation damage of the Schiff base in phosphoserine aminotransferase (structure E) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Dubnovitsky, A.P, Ravelli, R.B.G, Popov, A.N, Papageorgiou, A.C. | | Deposit date: | 2005-01-20 | | Release date: | 2005-05-19 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Strain Relief at the Active Site of Phosphoserine Aminotransferase Induced by Radiation Damage.
Protein Sci., 14, 2005
|
|
2BW1
 
 | | Iron-bound crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DPS-LIKE PEROXIDE RESISTANCE PROTEIN, ... | | Authors: | Kauko, A, Pulliainen, A, Haataja, S, Finne, J, Papageorgiou, A.C. | | Deposit date: | 2005-07-07 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Iron incorporation in Streptococcus suis Dps-like peroxide resistance protein Dpr requires mobility in the ferroxidase center and leads to the formation of a ferrihydrite-like core.
J. Mol. Biol., 364, 2006
|
|
1H0D
 
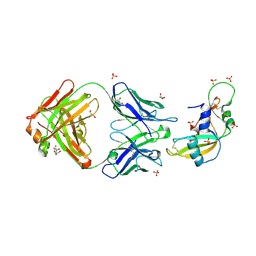 | | Crystal structure of Human Angiogenin in complex with Fab fragment of its monoclonal antibody mAb 26-2F | | Descriptor: | ANGIOGENIN, ANTIBODY FAB FRAGMENT, HEAVY CHAIN, ... | | Authors: | Chavali, G.B, Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 2002-06-19 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Angiogenin in Complex with an Antitumor Neutralizing Antibody
Structure, 11, 2003
|
|
4BYM
 
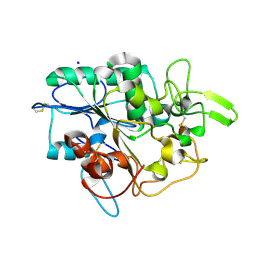 | | Structure of PhaZ7 PHB depolymerase Y105E mutant | | Descriptor: | CHLORIDE ION, PHB DEPOLYMERASE PHAZ7, SODIUM ION | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-07-20 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
4BTV
 
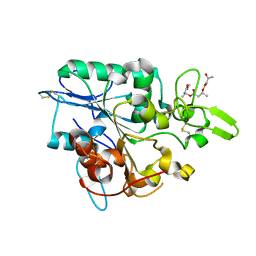 | | Structure of PhaZ7 PHB depolymerase in complex with 3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, PHB DEPOLYMERASE PHAZ7 | | Authors: | Hermawan, S, Subedi, B, Papageorgiou, A.C, Jendrossek, D. | | Deposit date: | 2013-06-19 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biochemical Analysis and Structure Determination of Paucimonas Lemoignei Poly(3-Hydroxybutyrate) (Phb) Depolymerase Phaz7 Muteins Reveal the Phb Binding Site and Details of Substrate-Enzyme Interactions.
Mol.Microbiol., 90, 2013
|
|
2C0R
 
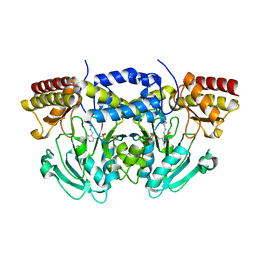 | |
2X76
 
 | | The crystal structure of PhaZ7 at atomic (1.2 Angstrom) resolution reveals details of the active site and suggests a substrate binding mode | | Descriptor: | CHLORIDE ION, GLYCEROL, IODIDE ION, ... | | Authors: | Wakadkar, S, Hermawan, S, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2010-02-24 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of PhaZ7 at atomic (1.2 A) resolution reveals details of the active site and suggests a substrate-binding mode.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 66, 2010
|
|
2X5X
 
 | | The crystal structure of PhaZ7 at atomic (1.2 Angstrom) resolution reveals details of the active site and suggests a substrate binding mode | | Descriptor: | CHLORIDE ION, IODIDE ION, PHB DEPOLYMERASE PHAZ7, ... | | Authors: | Wakadkar, S, Hermawan, S, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of PhaZ7 at atomic (1.2 A) resolution reveals details of the active site and suggests a substrate-binding mode.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 66, 2010
|
|
2YCD
 
 | | Structure of a novel Glutathione Transferase from Agrobacterium tumefaciens. | | Descriptor: | GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, S-(P-NITROBENZYL)GLUTATHIONE | | Authors: | Skopelitou, K, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2011-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Glutathione Transferase from Agrobacterium Tumefaciens Reveals a Novel Class of Bacterial Gst Superfamily.
Plos One, 7, 2012
|
|
2WLU
 
 | | Iron-bound crystal structure of Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, FE (III) ION, GLYCEROL, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-26 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
2WLA
 
 | | Streptococcus pyogenes Dpr | | Descriptor: | DPS-LIKE PEROXIDE RESISTANCE PROTEIN, GLYCEROL | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2009-06-23 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Streptococcus Pyogenes Dpr Reveal a Dodecameric Iron-Binding Protein with a Ferroxidase Site.
J.Biol.Inorg.Chem., 15, 2010
|
|
2WLT
 
 | |
2XKQ
 
 | | Crystal structure of Streptococcus suis Dpr with manganese | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Haikarainen, T, Thanassoulas, A, Stavros, P, Nounesis, G, Haataja, S, Papageorgiou, A.C. | | Deposit date: | 2010-07-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Thermodynamic Characterization of Metal Ion Binding in Streptococcus Suis Dpr.
J.Mol.Biol., 405, 2011
|
|
2VO4
 
 | | Glutathione transferase from Glycine max | | Descriptor: | 2,4-D INDUCIBLE GLUTATHIONE S-TRANSFERASE, 4-NITROPHENYL METHANETHIOL, GLYCEROL, ... | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and Functional Characterization of the Fluorodifen-Inducible Glutathione Transferase from Glycine Max Reveals an Active Site Topography Suited for Diphenylether Herbicides and a Novel L-Site.
J.Mol.Biol., 385, 2009
|
|
2XJN
 
 | | Crystal structure of Streptococcus suis Dpr with copper | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Haikarainen, T, Thanassoulas, A, Stavros, P, Nounesis, G, Haataja, S, Papageorgiou, A.C. | | Deposit date: | 2010-07-06 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Thermodynamic Characterization of Metal Ion Binding in Streptococcus Suis Dpr.
J.Mol.Biol., 405, 2011
|
|
2XJM
 
 | | Crystal structure of Streptococcus suis Dpr with cobalt | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Haikarainen, T, Thanassoulas, A, Stavros, P, Nounesis, G, Haataja, S, Papageorgiou, A.C. | | Deposit date: | 2010-07-06 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Thermodynamic Characterization of Metal Ion Binding in Streptococcus Suis Dpr.
J.Mol.Biol., 405, 2011
|
|
2WT4
 
 | |
2XGW
 
 | | ZINC-BOUND CRYSTAL STRUCTURE OF STREPTOCOCCUS PYOGENES DPR | | Descriptor: | CHLORIDE ION, GLYCEROL, PEROXIDE RESISTANCE PROTEIN, ... | | Authors: | Haikarainen, T, Tsou, C.-C, Wu, J.-J, Papageorgiou, A.C. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization and Biological Implications of Di-Zinc Binding in the Ferroxidase Center of Streptococcus Pyogenes Dpr.
Biochem.Biophys.Res.Commun., 398, 2010
|
|
