1DUA
 
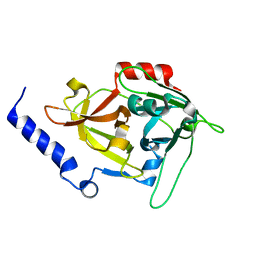 | |
1B1Z
 
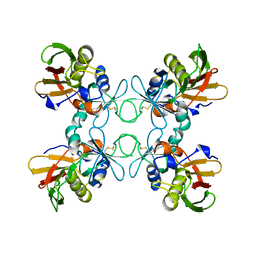 | | STREPTOCOCCAL PYROGENIC EXOTOXIN A1 | | Descriptor: | PROTEIN (TOXIN) | | Authors: | Papageorgiou, A.C, Acharya, K.R. | | Deposit date: | 1998-11-24 | | Release date: | 1999-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for the recognition of superantigen streptococcal pyrogenic exotoxin A (SpeA1) by MHC class II molecules and T-cell receptors.
EMBO J., 18, 1999
|
|
4TOP
 
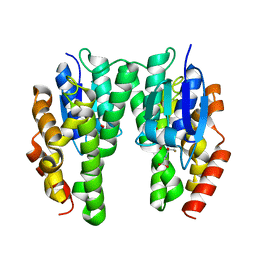 | | Glycine max glutathione transferase | | Descriptor: | 2,4-D inducible glutathione S-transferase, GLUTATHIONE | | Authors: | Axarli, I, Dhavala, P, Papageorgiou, A.C. | | Deposit date: | 2014-06-06 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Comparative analysis of the structural and functional features of two homologous tau class glutathione transferases from Glycine max
To Be Published
|
|
8APP
 
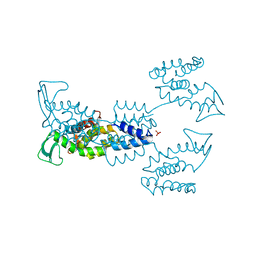 | | AbLys1 endolysin from Acinetobacter baumannii phage AbTZA1 | | Descriptor: | Endolysin, GLYCEROL, PHOSPHATE ION | | Authors: | Premetis, G.E, Stathi, A, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2022-08-10 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Characterization of a glycoside hydrolase endolysin from Acinetobacter baumannii phage AbTZA1 with high antibacterial potency and novel structural features.
Febs J., 290, 2023
|
|
4ZU2
 
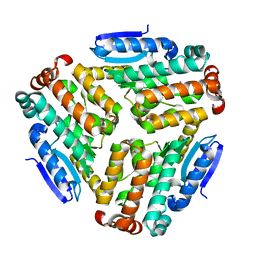 | | Pseudomonas aeruginosa AtuE | | Descriptor: | IODIDE ION, Putative isohexenylglutaconyl-CoA hydratase | | Authors: | Poudel, N, Pfannstiel, J, Simon, O, Walter, N, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2015-05-15 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Pseudomonas aeruginosa Isohexenyl Glutaconyl Coenzyme A Hydratase (AtuE) Is Upregulated in Citronellate-Grown Cells and Belongs to the Crotonase Family.
Appl.Environ.Microbiol., 81, 2015
|
|
5AGY
 
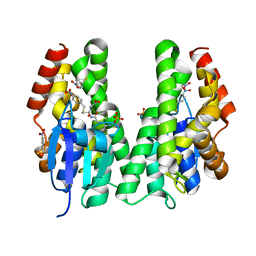 | | CRYSTAL STRUCTURE OF A TAU CLASS GST MUTANT FROM GLYCINE | | Descriptor: | 4-NITROPHENYL METHANETHIOL, GLUTATHIONE S-TRANSFERASE, PHOSPHATE ION, ... | | Authors: | Axarli, I, Muleta, A.W, Vlachakis, D, Kossida, S, Kotzia, G, Dhavala, P, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2015-02-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Directed Evolution of Tau Class Glutathione Transferases Reveals a Site that Regulates Catalytic Efficiency and Masks Cooperativity.
Biochem.J., 473, 2016
|
|
5LHF
 
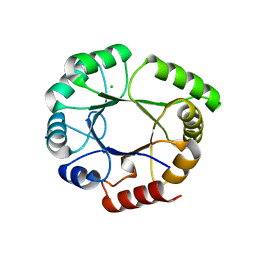 | |
5LCZ
 
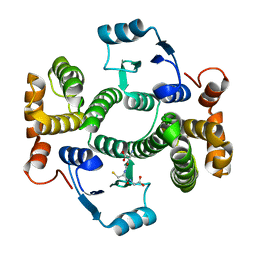 | | Chimeric GST | | Descriptor: | GLUTATHIONE, Glutathione S-transferase A1,Glutathione S-transferase alpha-2,Glutathione S-transferase A1,Glutathione S-transferase alpha-2,Glutathione S-transferase A1 | | Authors: | Axarli, A, Muleta, A.W, Chronopoulou, E.G, Papageorgiou, A.C, Labrou, N.E. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.325 Å) | | Cite: | Directed evolution of glutathione transferases towards a selective glutathione-binding site and improved oxidative stability.
Biochim. Biophys. Acta, 1861, 2017
|
|
5LHE
 
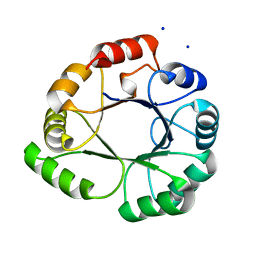 | |
1QIL
 
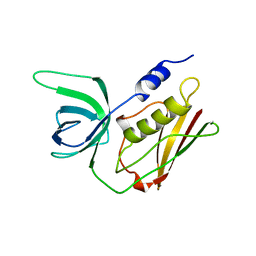 | |
1ABB
 
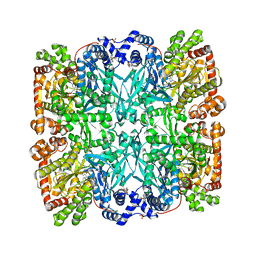 | | CONTROL OF PHOSPHORYLASE B CONFORMATION BY A MODIFIED COFACTOR: CRYSTALLOGRAPHIC STUDIES ON R-STATE GLYCOGEN PHOSPHORYLASE RECONSTITUTED WITH PYRIDOXAL 5'-DIPHOSPHATE | | Descriptor: | GLYCOGEN PHOSPHORYLASE B, INOSINIC ACID, PYRIDOXAL-5'-DIPHOSPHATE, ... | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Papageorgiou, A.C, Acharya, K.R, Barford, D, Johnson, L.N. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of phosphorylase b conformation by a modified cofactor: crystallographic studies on R-state glycogen phosphorylase reconstituted with pyridoxal 5'-diphosphate.
Protein Sci., 1, 1992
|
|
1PHK
 
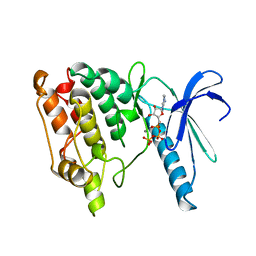 | | TWO STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHORYLASE, KINASE: AN ACTIVE PROTEIN KINASE COMPLEXED WITH NUCLEOTIDE, SUBSTRATE-ANALOGUE AND PRODUCT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PHOSPHORYLASE KINASE | | Authors: | Owen, D.J, Noble, M.E.M, Garman, E.F, Papageorgiou, A.C, Johnson, L.N. | | Deposit date: | 1996-03-15 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product.
Structure, 3, 1995
|
|
6YEU
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
6YET
 
 | | Second EH domain of AtEH1/Pan1 | | Descriptor: | CALCIUM ION, Calcium-binding EF hand family protein | | Authors: | Yperman, K, Papageorgiou, A, Evangelidis, T, Van Damme, D, Tripsianes, K. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding.
Nat Commun, 12, 2021
|
|
2J4X
 
 | | Streptococcus dysgalactiae-derived mitogen (SDM) | | Descriptor: | GLYCEROL, MITOGEN, ZINC ION | | Authors: | Saarinen, S, Kato, H, Uchiyama, T, Miyoshi-Akiyama, T, Papageorgiou, A.C. | | Deposit date: | 2006-09-07 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Streptococcus Dysgalactiae-Derived Mitogen Reveals a Zinc-Binding Site and Alterations in Tcr Binding.
J.Mol.Biol., 373, 2007
|
|
7Q1K
 
 | | Crystal structure of the native AA9A LPMO from Thermoascus aurantiacus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Yu, W, Mohsin, I, Li, D.C, Papageorgiou, A.C. | | Deposit date: | 2021-10-20 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Purification and Structural Characterization of the Auxiliary Activity 9 Native Lytic Polysaccharide Monooxygenase from Thermoascus aurantiacus and Identification of Its C1- and C4-Oxidized Reaction Products
Catalysts, 12, 2022
|
|
7OX6
 
 | | Solution structure of human interleukin-9 | | Descriptor: | Interleukin-9 | | Authors: | Savvides, S.N, Tripsianes, K, De Vos, T, Papageorgiou, A, Evangelidis, T. | | Deposit date: | 2021-06-22 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-11 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mechanism and antagonism of receptor signaling mediated by Interleukin-9 (IL-9)
Biorxiv, 2022
|
|
5MLX
 
 | | Open loop conformation of PhaZ7 Y105E mutant | | Descriptor: | CHLORIDE ION, PHB depolymerase PhaZ7, SODIUM ION | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
5MLY
 
 | | Closed loop conformation of PhaZ7 Y105E mutant | | Descriptor: | PHB depolymerase PhaZ7 | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
1STE
 
 | |
6SZ6
 
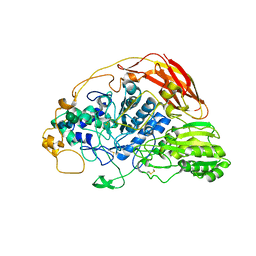 | | Chaetomium thermophilum beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Mohsin, I, Poudel, N, Papageorgiou, A.C. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.988 Å) | | Cite: | Crystal Structure of a GH3 beta-Glucosidase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 20, 2019
|
|
1W3U
 
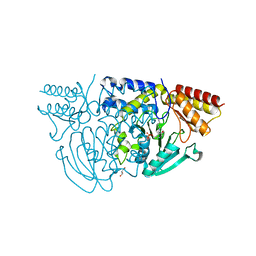 | |
1W23
 
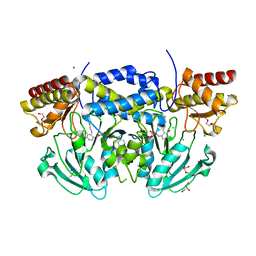 | | Crystal structure of phosphoserine aminotransferase from Bacillus alcalophilus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dubnovitsky, A, Kapetaniou, E.G, Papageorgiou, A.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Enzyme Adaptation to Alkaline Ph: Atomic Resolution (1.08 A) Structure of Phosphoserine Aminotransferase from Bacillus Alcalophilus
Protein Sci., 14, 2005
|
|
2QIL
 
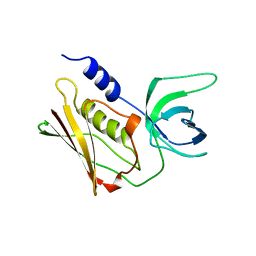 | |
1UMN
 
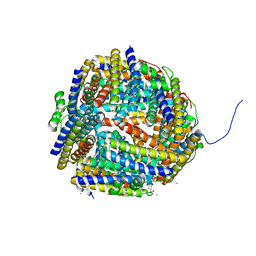 | | Crystal structure of Dps-like peroxide resistance protein (Dpr) from Streptococcus suis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kauko, A, Haataja, S, Pulliainen, A, Finne, J, Papageorgiou, A.C. | | Deposit date: | 2003-08-26 | | Release date: | 2004-04-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Streptococcus suis Dps-like peroxide resistance protein Dpr: implications for iron incorporation.
J. Mol. Biol., 338, 2004
|
|
