6B6B
 
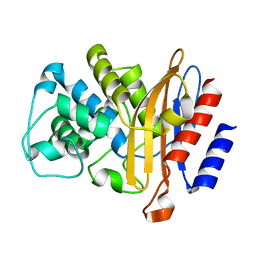 | |
6B6A
 
 | | Beta-Lactamase, 2secs timepoint, mixed, shards crystal form | | Descriptor: | (2R)-2-[(1S)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}-2-hydroxyethyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, Ceftriaxone, ... | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-06-27 | | Last modified: | 2020-01-15 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Enzyme intermediates captured "on the fly" by mix-and-inject serial crystallography.
BMC Biol., 16, 2018
|
|
4JTB
 
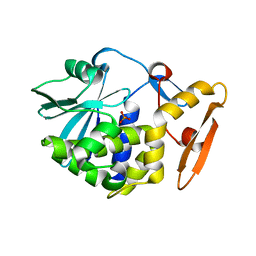 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, rRNA N-glycosidase | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Kushwaha, G.S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-23 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with phosphate ion at 1.71 Angstrom resolution
To be published
|
|
4JTP
 
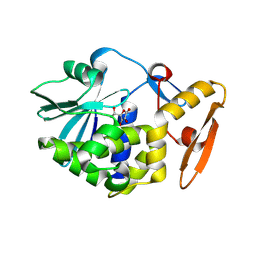 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Ascorbic acid at 1.85 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ASCORBIC ACID, rRNA N-glycosidase | | Authors: | Pandey, S, Bhushan, A, Singh, A, Tyagi, T.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-24 | | Release date: | 2013-04-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Ascorbic acid at 1.85 Angstrom resolution
TO BE PUBLISHED
|
|
4KL4
 
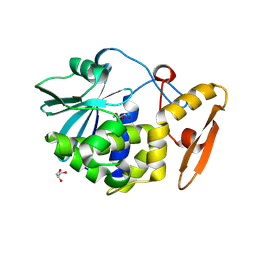 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pandey, S, Tyagi, T.K, Singh, A, Bhushan, A, Kushwaha, G.S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-05-07 | | Release date: | 2013-05-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina complexed with Polyethylene glycol at 1.90 Angstrom resolution
To be Published
|
|
5DVI
 
 | | High resolution crystal Structure of glucose complexed periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, GLYCEROL, SULFATE ION, ... | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
5DVF
 
 | | Crystal structure of unliganded periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, SULFATE ION | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
5DVJ
 
 | | Crystal structure of galactose complexed periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, GLYCEROL, SULFATE ION, ... | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
7FHZ
 
 | |
7FI1
 
 | |
7FI2
 
 | |
7FI0
 
 | | Crystal structure of Multi-functional Polysaccharide lyase Smlt1473 (WT) from Stenotrophomonas maltophilia (strain K279a) in ManA bound form at pH-5.0 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Polysaccharide lyase, SULFATE ION, ... | | Authors: | Pandey, S, Berger, B.W, Acharya, R. | | Deposit date: | 2021-07-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural insights into the mechanism of pH-selective substrate specificity of the polysaccharide lyase Smlt1473.
J.Biol.Chem., 297, 2021
|
|
7FHY
 
 | |
7FHX
 
 | |
7FHU
 
 | |
7FHV
 
 | |
7FHW
 
 | |
7AZT
 
 | | X-ray crystallographic structure of (6-4)photolyase from Drosophila melanogaster at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, RE11660p | | Authors: | Cellini, A, Wahlgren, W.Y, Henry, L, Westenhoff, S, Pandey, S. | | Deposit date: | 2020-11-17 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The three-dimensional structure of Drosophila melanogaster (6-4) photolyase at room temperature.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4XY7
 
 | | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with N-acetylglucosamine at 2.5 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein | | Authors: | Yamini, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the complex of ribosome inactivating protein from Momordica balsamina with N- acetylglucosamine at 2.5 A resolution
To Be Published
|
|
5ILW
 
 | | Crystal structure of the complex of type 1 Ribosome inactivating protein from Momordica balsamina with Uridine at 1.97 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
5ILX
 
 | | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, ... | | Authors: | Singh, P.K, Singh, A, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-03-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Ribosome inactivating protein from Momordica balsamina with Uracil at 1.70 Angstrom resolution
To Be Published
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
4ZU0
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine monophosphate at 1.80 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
4ZZ6
 
 | | Structure of the complex of type 1 ribosome inactivating protein from Momordica balsamina with a nucleotide, cytidine triphosphate at 2.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Yamin, S, Pandey, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein fromMomordica balsaminawith cytosine, cytidine, and cytidine diphosphate.
Biochem Biophys Rep, 4, 2015
|
|
6T3L
 
 | | PAS-GAF fragment from Deinococcus radiodurans phytochrome in dark state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Claesson, E, Takala, H, Yuan Wahlgren, W, Pandey, S, Schmidt, M, Westenhoff, S. | | Deposit date: | 2019-10-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The primary structural photoresponse of phytochrome proteins captured by a femtosecond X-ray laser.
Elife, 9, 2020
|
|
