3RFN
 
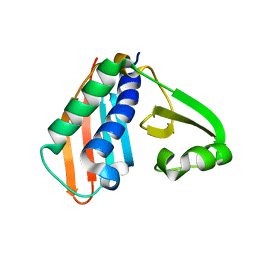 | | Epitope backbone grafting by computational design for improved presentation of linear epitopes on scaffold proteins | | Descriptor: | BB_1wnu_001, ZINC ION | | Authors: | Azoitei, M.L, Ban, Y.A, Julien, J.P, Bryson, S, Schroeter, A, Kalyuzhniy, O, Porter, J.R, Adachi, Y, Baker, D, Szabo, E, Pai, E.F, Schief, W.R. | | Deposit date: | 2011-04-06 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope.
J.Mol.Biol., 415, 2012
|
|
3SZE
 
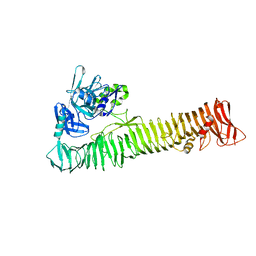 | | Crystal structure of the passenger domain of the E. coli autotransporter EspP | | Descriptor: | Serine protease espP | | Authors: | Khan, S, Mian, H.S, Sandercock, L.E, Battaile, K.P, Lam, R, Chirgadze, N.Y, Pai, E.F. | | Deposit date: | 2011-07-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
J.Mol.Biol., 413, 2011
|
|
3R3Z
 
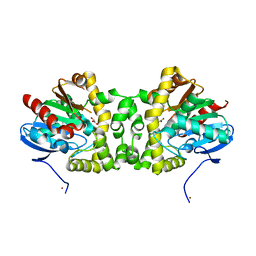 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/Glycolate | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID, NICKEL (II) ION | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3R3Y
 
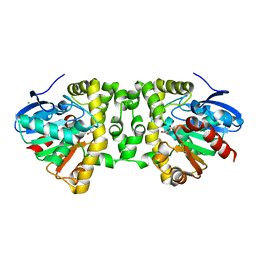 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
3SW6
 
 | |
3THQ
 
 | |
3R3W
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fluoroacetate dehalogenase, ... | | Authors: | Chan, P.W.Y, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-03-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the reaction coordinates of enzymatic defluorination.
J.Am.Chem.Soc., 133, 2011
|
|
121P
 
 | | STRUKTUR UND GUANOSINTRIPHOSPHAT-HYDROLYSEMECHANISMUS DES C-TERMINAL VERKUERZTEN MENSCHLICHEN KREBSPROTEINS P21-H-RAS | | Descriptor: | H-RAS P21 PROTEIN, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Krengel, U, Scheffzek, K, Scherer, A, Kabsch, W, Wittinghofer, A, Pai, E.F. | | Deposit date: | 1991-06-06 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Struktur Und Guanosintriphosphat-Hydrolysemechanismus Des C-Terminal Verkuerzten Menschlichen Krebsproteins P21-H-Ras
Thesis, 1991
|
|
1AVT
 
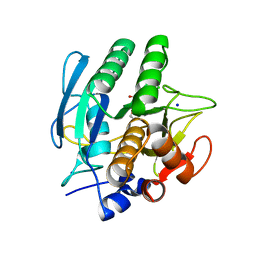 | | SUBTILISIN CARLSBERG D-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-19 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1AV7
 
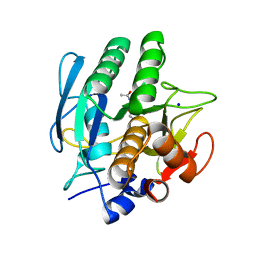 | | SUBTILISIN CARLSBERG L-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-29 | | Release date: | 1998-04-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1DV7
 
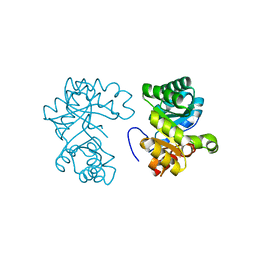 | |
1DVJ
 
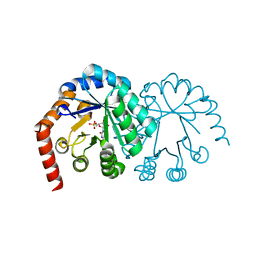 | | CRYSTAL STRUCTURE OF OROTIDINE MONOPHOSPHATE DECARBOXYLASE COMPLEXED WITH 6-AZAUMP | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, OROTIDINE 5'-PHOSPHATE DECARBOXYLASE | | Authors: | Wu, N, Mo, Y, Gao, J, Pai, E.F. | | Deposit date: | 2000-03-30 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Electrostatic stress in catalysis: structure and mechanism of the enzyme orotidine monophosphate decarboxylase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EJ2
 
 | | Crystal structure of methanobacterium thermoautotrophicum nicotinamide mononucleotide adenylyltransferase with bound NAD+ | | Descriptor: | NICOTINAMIDE MONONUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Saridakis, V, Christendat, D, Kimber, M.S, Edwards, A.M, Pai, E.F, Midwest Center for Structural Genomics (MCSG), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-29 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.
J.Biol.Chem., 276, 2001
|
|
1KM2
 
 | |
1KM0
 
 | |
1KM1
 
 | |
1KLZ
 
 | |
1KM6
 
 | |
1KM3
 
 | |
1LOS
 
 | |
1LOQ
 
 | |
1LP6
 
 | |
1LOL
 
 | | Crystal structure of orotidine monophosphate decarboxylase complex with XMP | | Descriptor: | 1,3-BUTANEDIOL, XANTHOSINE-5'-MONOPHOSPHATE, orotidine 5'-monophosphate decarboxylase | | Authors: | Wu, N, Pai, E.F. | | Deposit date: | 2002-05-06 | | Release date: | 2002-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of inhibitor complexes reveal an alternate binding mode in orotidine-5'-monophosphate decarboxylase.
J.Biol.Chem., 277, 2002
|
|
