6DNO
 
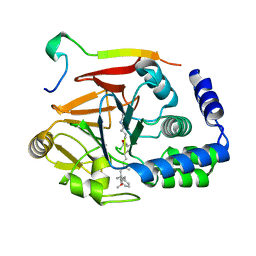 | | Crystal structure of Protein Phosphatase 1 (PP1) bound to the muscle glycogen-targeting subunit (Gm) | | Descriptor: | Microcystin-LR, Protein phosphatase 1 regulatory subunit 3A, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Identification of the substrate recruitment mechanism of the muscle glycogen protein phosphatase 1 holoenzyme.
Sci Adv, 4, 2018
|
|
6BSR
 
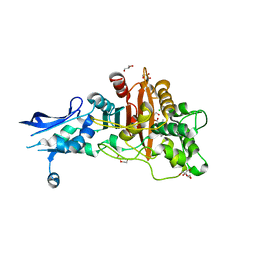 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6BSQ
 
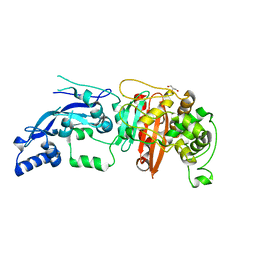 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | Descriptor: | CHLORIDE ION, GLYCEROL, PBP4 protein | | Authors: | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2017-12-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKY
 
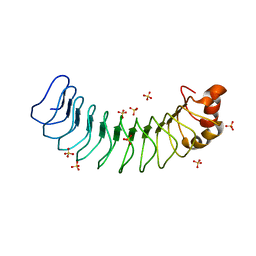 | | Human SDS22 | | Descriptor: | Protein phosphatase 1 regulatory subunit 7, SULFATE ION | | Authors: | Choy, M.S, Bolik-Coulon, N, Page, R, Peti, W. | | Deposit date: | 2018-09-26 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of SDS22 provides insights into the mechanism of heterodimer formation with PP1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6MKI
 
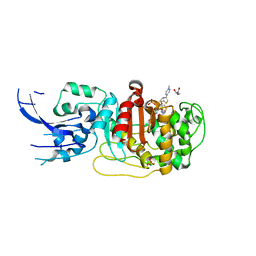 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the ceftaroline-bound form | | Descriptor: | Ceftaroline, bound form, GLYCEROL, ... | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6MKJ
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the closed conformation | | Descriptor: | penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Soares, A, D'Andrea, E.D, Jaconcic, J, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
