5RJI
 
 | | PanDDA analysis group deposition of ground-state model of PHIP | | Descriptor: | PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJX
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z285782452 | | Descriptor: | N-methyl-2-(methylsulfonyl)aniline, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKD
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z2168282707 | | Descriptor: | (6S)-1-methyl-4,5,6,7-tetrahydro-1H-benzotriazole-6-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.237 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKU
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z32367954 | | Descriptor: | PH-interacting protein, ~{N}-cyclopropyl-1,3-benzodioxole-5-carboxamide | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5V2D
 
 | |
5RJK
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024661 | | Descriptor: | 5-bromo-2-hydroxybenzonitrile, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK0
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z30932204 | | Descriptor: | N-[(4-sulfamoylphenyl)methyl]acetamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKG
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1124201124 | | Descriptor: | PH-interacting protein, ethyl 1~{H}-pyrazole-4-carboxylate | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.282 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKW
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z27678561 | | Descriptor: | N-(6-methoxy-1,3-benzothiazol-2-yl)cyclopropanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00023833 | | Descriptor: | 4-bromanyl-1,8-naphthyridine, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJY
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z383325512 | | Descriptor: | N-[(1H-pyrazol-4-yl)methyl]acetamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKE
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z906021418 | | Descriptor: | 5-chloro-2-(propan-2-yl)pyrimidine-4-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKS
 
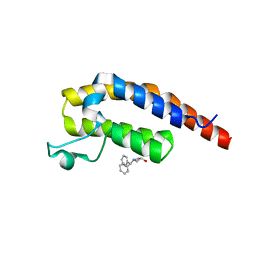 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1696844792 | | Descriptor: | 1-(diphenylmethyl)azetidin-3-ol, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5SXX
 
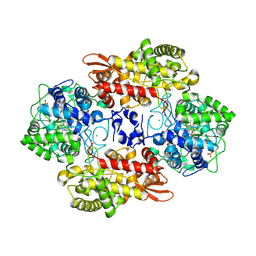 | |
5SYU
 
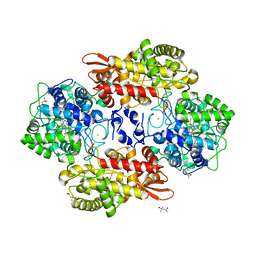 | | Crystal structure of Burkholderia pseudomallei KatG E242Q variant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-12 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalase Activity of Catalase-Peroxidases Is Modulated by Changes in the pKa of the Distal Histidine.
Biochemistry, 56, 2017
|
|
5SXR
 
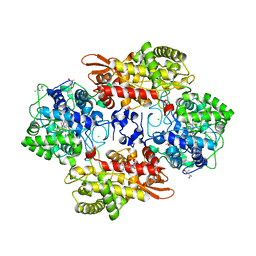 | | Crystal structure of B. pseudomallei KatG with NAD bound | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases.
J. Biol. Chem., 285, 2010
|
|
5SXQ
 
 | |
5SYH
 
 | | Structure of D141A variant of B. pseudomallei KatG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An ionizable active-site tryptophan imparts catalase activity to a peroxidase core.
J. Am. Chem. Soc., 136, 2014
|
|
5SYI
 
 | | Structure of D141A variant of B. pseudomallei KatG complexed with INH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitubercular pro-drug isoniazid in the heme access channel of catalase-peroxidase (KatG). A combined structural and metadynamics investigation.
J Phys Chem B, 118, 2014
|
|
3ZCH
 
 | | Ascorbate peroxidase W41A-H42M mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of Ascorbate Peroxidase
Dalton Trans, 42, 2013
|
|
3RNJ
 
 | | Crystal structure of the SH3 domain from IRSp53 (BAIAP2) | | Descriptor: | 1,2-ETHANEDIOL, Brain-specific angiogenesis inhibitor 1-associated protein 2, ISOPROPYL ALCOHOL, ... | | Authors: | Simister, P.C, Barilari, M, Muniz, J.R.C, Dente, L, Knapp, S, von Delft, F, Filippakopoulos, P, Vollmar, M, Chaikuad, A, Raynor, J, Tregubova, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the SH3 domain from IRSp53 (BAIAP2)
To be Published
|
|
3ZCY
 
 | | Ascorbate peroxidase W41A-H42Y mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASCORBATE PEROXIDASE, POTASSIUM ION, ... | | Authors: | Gumiero, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2012-11-23 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Conformational Mobility of the Active Site of Ascorbate Peroxidase
Dalton Trans, 42, 2013
|
|
5RJU
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z57261895 | | Descriptor: | 2-cyclohexyl-N-(4H-1,2,4-triazol-4-yl)acetamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKA
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z126932614 | | Descriptor: | 2-[(methylsulfonyl)methyl]-1H-benzimidazole, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKO
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z30620520 | | Descriptor: | PH-interacting protein, cyclopropyl-[4-(4-fluorophenyl)piperazin-1-yl]methanone | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
