1QKJ
 
 | | T4 Phage B-Glucosyltransferase, Substrate Binding and Proposed Catalytic Mechanism | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Imberty, I, Aschke-Sonnenborn, U, Ruger, W, Freemont, P.S. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | T4 Phage Beta-Glucosyltransferase: Substrate Binding and Proposed Catalytic Mechanism
J.Mol.Biol., 292, 1999
|
|
4PHG
 
 | | Crystal structure of Ypt7 covalently modified with GTP | | Descriptor: | GTP-binding protein YPT7, MAGNESIUM ION, N-[3-(propanoylamino)propyl]guanosine 5'-(tetrahydrogen triphosphate), ... | | Authors: | Koch, D, Wiegandt, D, Vieweg, S, Hofmann, F, Wu, Y, Itzen, A, Mueller, M.P, Goody, R.S. | | Deposit date: | 2014-05-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Locking GTPases covalently in their functional states.
Nat Commun, 6, 2015
|
|
2XO1
 
 | | xpt-pbuX C74U Riboswitch from B. subtilis bound to N6-methyladenine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), Guanine riboswitch, ... | | Authors: | Daldrop, P, Reyes, F.E, Robinson, D.A, Hammond, C.M, Lilley, D.M.J, Batey, R.T, Brenk, R. | | Deposit date: | 2010-08-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel ligands for a purine riboswitch discovered by RNA-ligand docking.
Chem. Biol., 18, 2011
|
|
7UC3
 
 | |
4GM5
 
 | | Carboxypeptidase T with Sulphamoil Arginine | | Descriptor: | CALCIUM ION, Carboxypeptidase T, GLYCEROL, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2012-08-15 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Carboxypeptidase T with Sulphamoil Arginine
To be Published
|
|
2BCW
 
 | | Coordinates of the N-terminal domain of ribosomal protein L11,C-terminal domain of ribosomal protein L7/L12 and a portion of the G' domain of elongation factor G, as fitted into cryo-em map of an Escherichia coli 70S*EF-G*GDP*fusidic acid complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L7/L12, Elongation factor G | | Authors: | Datta, P.P, Sharma, M.R, Qi, L, Frank, J, Agrawal, R.K. | | Deposit date: | 2005-10-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.2 Å) | | Cite: | Interaction of the G' Domain of Elongation Factor G and the C-Terminal Domain of Ribosomal Protein L7/L12 during Translocation as Revealed by Cryo-EM.
Mol.Cell, 20, 2005
|
|
2O1Z
 
 | | Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155) | | Descriptor: | FE (III) ION, Ribonucleotide Reductase Subunit R2, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Tempel, W, Qiu, W, Lew, J, Wernimont, A.K, Lin, Y.H, Hassanali, A, Melone, M, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
To be Published
|
|
4PBT
 
 | | Crystal structure of the M. jannaschii G2 tRNA synthetase variant bound to 4-trans-cyclooctene-amidopheylalanine (Tco-amF) | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 4-{[(1R,4E)-cyclooct-4-en-1-ylcarbonyl]amino}-L-phenylalanine, GLYCEROL, ... | | Authors: | Cooley, R.B, Karplus, P.A, Mehl, R.A. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gleaning Unexpected Fruits from Hard-Won Synthetases: Probing Principles of Permissivity in Non-canonical Amino Acid-tRNA Synthetases.
Chembiochem, 15, 2014
|
|
4PCR
 
 | | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) complexed with Gamma-Aminobutyric Acid (GABA) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Concanavalin-Br, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Silva-Filho, J.C, Teixeira, C.S, Cavada, B.S, Nascimento, K.S, Nbrega, R.B, Nagano, C.S, Sampaio, A.H, Leal, R.B, Neto, I.L.B. | | Deposit date: | 2014-04-16 | | Release date: | 2015-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Canavalia brasiliensis seed lectin (ConBr) complexed with Gamma-Aminobutyric Acid (GABA)
To Be Published
|
|
1NPO
 
 | |
4FU3
 
 | | CID of human RPRD1B | | Descriptor: | CHLORIDE ION, Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CID of human RPRD1B
TO BE PUBLISHED
|
|
2F1K
 
 | | Crystal structure of Synechocystis arogenate dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, prephenate dehydrogenase | | Authors: | Legrand, P, Dumas, R, Seux, M, Rippert, P, Ravelli, R, Ferrer, J.-L, Matringe, M. | | Deposit date: | 2005-11-14 | | Release date: | 2006-05-09 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biochemical Characterization and Crystal Structure of Synechocystis Arogenate Dehydrogenase Provide Insights into Catalytic Reaction
Structure, 14, 2006
|
|
1OJ6
 
 | | Human brain neuroglobin three-dimensional structure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Nardini, M, Moens, L, Ascenzi, P, Hankeln, T, Burmester, T, Bolognesi, M. | | Deposit date: | 2003-07-03 | | Release date: | 2003-09-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human Brain Neuroglobin Structure Reveals a Distinct Mode of Controlling Oxygen Affinity
Structure, 11, 2003
|
|
2BDG
 
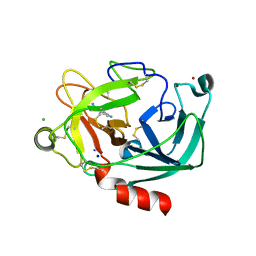 | | Human Kallikrein 4 complex with nickel and p-aminobenzamidine | | Descriptor: | CHLORIDE ION, Kallikrein-4, NICKEL (II) ION, ... | | Authors: | Debela, M, Bode, W, Goettig, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-10-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of human tissue kallikrein 4: activity modulation by a specific zinc binding site.
J.Mol.Biol., 362, 2006
|
|
2Y6M
 
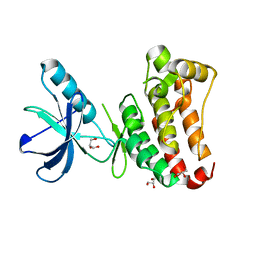 | |
1Q9K
 
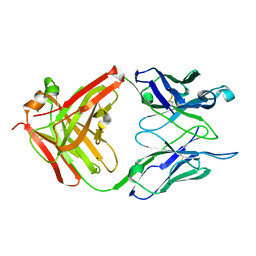 | | S25-2 Fab Unliganded 1 | | Descriptor: | MAGNESIUM ION, S25-2 Fab (IgG1k) heavy chain, S25-2 Fab (IgG1k) light chain | | Authors: | Nguyen, H.P, Seto, N.O, MacKenzie, C.R, Brade, L, Kosma, P, Brade, H, Evans, S.V. | | Deposit date: | 2003-08-25 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Germline antibody recognition of distinct carbohydrate epitopes.
Nat.Struct.Biol., 10, 2003
|
|
2YAA
 
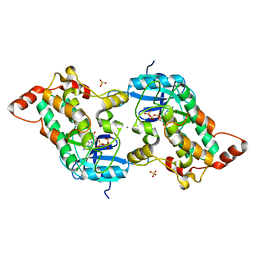 | |
7BHS
 
 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
4N2C
 
 | | Crystal structure of Protein Arginine Deiminase 2 (F221/222A, 10 mM Ca2+) | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.022 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
1QC0
 
 | | CRYSTAL STRUCTURE OF A 19 BASE PAIR COPY CONTROL RELATED RNA DUPLEX | | Descriptor: | 5'-R(*GP*CP*AP*CP*CP*GP*CP*UP*AP*CP*CP*AP*AP*CP*GP*GP*UP*GP*C)-3', 5'-R(*GP*CP*AP*CP*CP*GP*UP*UP*GP*GP*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', 5'-R(*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', ... | | Authors: | Klosterman, P.S, Shah, S.A, Steitz, T.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two plasmid copy control related RNA duplexes: An 18 base pair duplex at 1.20 A resolution and a 19 base pair duplex at 1.55 A resolution.
Biochemistry, 38, 1999
|
|
7UI8
 
 | |
4G0L
 
 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
3TBS
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX THE WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCOPROTEIN G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
1YCR
 
 | |
2BYS
 
 | | CRYSTAL STRUCTURE OF ACHBP FROM APLYSIA CALIFORNICA IN complex with lobeline | | Descriptor: | ACETYLCHOLINE-BINDING PROTEIN, LOBELINE | | Authors: | Hansen, S.B, Sulzenbacher, G, Huxford, T, Marchot, P, Taylor, P, Bourne, Y. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Aplysia Achbp Complexes with Nicotinic Agonists and Antagonists Reveal Distinctive Binding Interfaces and Conformations.
Embo J., 24, 2005
|
|
