3FRW
 
 | | Crystal structure of putative TrpR protein from Ruminococcus obeum | | Descriptor: | ACETATE ION, Putative Trp repressor protein | | Authors: | Osipiuk, J, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-20 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TrpR protein from Ruminococcus obeum.
To be Published
|
|
3PGY
 
 | | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCINE, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant
To be Published
|
|
3GHZ
 
 | | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Salmonella typhimurium | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Gu, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | X-ray crystal structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Salmonella typhimurium.
To be Published
|
|
3CZQ
 
 | | Crystal structure of putative polyphosphate kinase 2 from Sinorhizobium meliloti | | Descriptor: | FORMIC ACID, GLYCEROL, Putative polyphosphate kinase 2 | | Authors: | Osipiuk, J, Evdokimova, E, Nocek, B, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3Q12
 
 | | Pantoate-beta-alanine ligase from Yersinia pestis in complex with pantoate. | | Descriptor: | CHLORIDE ION, PANTOATE, Pantoate--beta-alanine ligase | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-16 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Pantoate-beta-alanine ligase from Yersinia pestis.
To be Published
|
|
3QU1
 
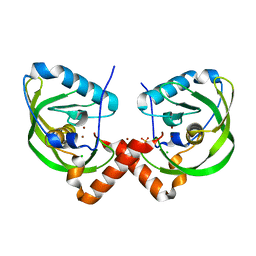 | | Peptide deformylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Peptide deformylase 2, SULFATE ION, ... | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide deformylase from Vibrio cholerae.
To be Published
|
|
3QWU
 
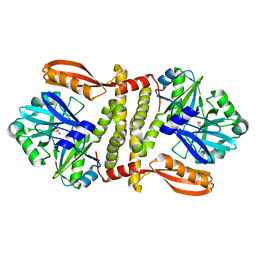 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
3Q6D
 
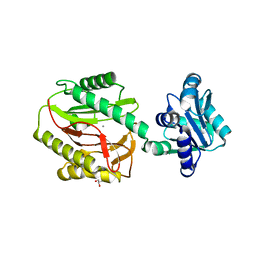 | | Xaa-Pro dipeptidase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, GLYCEROL, Proline dipeptidase | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Xaa-Pro dipeptidase from Bacillus anthracis.
To be Published
|
|
3C2B
 
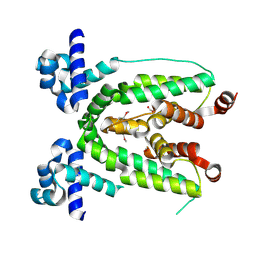 | | Crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | FORMIC ACID, Transcriptional regulator, TetR family | | Authors: | Osipiuk, J, Skarina, T, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens.
To be Published
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3BED
 
 | |
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CWC
 
 | | Crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative glycerate kinase 2 | | Authors: | Osipiuk, J, Zhou, M, Holzle, D, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-08 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | X-ray crystal structure of putative glycerate kinase 2 from Salmonella typhimurium LT2.
To be Published
|
|
3DQG
 
 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | Descriptor: | Heat shock 70 kDa protein F | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
3CTV
 
 | | Crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-14 | | Release date: | 2008-04-29 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | X-ray crystal structure of central domain of 3-hydroxyacyl-CoA dehydrogenase from Archaeoglobus fulgidus.
To be Published
|
|
3DT5
 
 | | C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Uncharacterized protein AF_0924 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-14 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray crystal structure of C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus.
To be Published
|
|
3DNX
 
 | |
3BRU
 
 | |
3DV9
 
 | | Putative beta-phosphoglucomutase from Bacteroides vulgatus. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-18 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of putative beta-phosphoglucomutase from Bacteroides vulgatus.
To be Published
|
|
3DOB
 
 | | Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans. | | Descriptor: | BETA-MERCAPTOETHANOL, Heat shock 70 kDa protein F44E5.5 | | Authors: | Osipiuk, J, Hatzos, C, Gu, M, Zhang, R, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-03 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | X-ray crystal structure of Peptide-binding domain of Heat shock 70 kDa protein F44E5.5 from C.elegans.
To be Published
|
|
3E8X
 
 | | Putative NAD-dependent epimerase/dehydratase from Bacillus halodurans. | | Descriptor: | CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative NAD-dependent epimerase/dehydratase | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-20 | | Release date: | 2008-09-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of putative NAD-dependent epimerase/dehydratase from Bacillus halodurans.
To be Published
|
|
3O6C
 
 | | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate synthase | | Authors: | Osipiuk, J, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Pyridoxal phosphate biosynthetic protein PdxJ from Campylobacter jejuni.
To be Published
|
|
3H0X
 
 | | Crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae | | Descriptor: | 78 kDa glucose-regulated protein homolog | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Sahi, C, Craig, E.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of Kar2 protein from Saccharomyces cerevisiae.
To be Published
|
|
3GO9
 
 | | Predicted insulinase family protease from Yersinia pestis | | Descriptor: | DI(HYDROXYETHYL)ETHER, ZINC ION, insulinase family protease | | Authors: | Osipiuk, J, Maltseva, N, Onopriyenko, O, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structure of predicted insulinase family protease from Yersinia pestis.
To be Published
|
|
3P4E
 
 | | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Osipiuk, J, Zhou, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Phosphoribosylformylglycinamidine cyclo-ligase from Vibrio cholerae.
To be Published
|
|
