3X1O
 
 | | Crystal structure of the ROQ domain of human Roquin | | Descriptor: | IODIDE ION, Roquin-1 | | Authors: | Ose, T, Verma, A, Cockburn, J.B, Berrow, N.S, Alderton, D, Stuart, D, Owens, R.J, Jones, E.Y. | | Deposit date: | 2014-11-26 | | Release date: | 2015-03-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis
Nat Commun, 6, 2015
|
|
2N37
 
 | | Solution structure of AVR-Pia | | Descriptor: | AVR-Pia protein | | Authors: | Ose, T, Oikawa, A, Nakamura, Y, Maenaka, K, Higuchi, Y, Satoh, Y, Fujiwara, S, Demura, M, Sone, T. | | Deposit date: | 2015-05-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an avirulence protein, AVR-Pia, from Magnaporthe oryzae
J.Biomol.Nmr, 63, 2015
|
|
1VCI
 
 | | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3 complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1V43
 
 | | Crystal Structure of ATPase subunit of ABC Sugar Transporter | | Descriptor: | sugar-binding transport ATP-binding protein | | Authors: | Ose, T, Fujie, T, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-11-08 | | Release date: | 2004-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the ATP-binding cassette of multisugar transporter from Pyrococcus horikoshii OT3
Proteins, 57, 2004
|
|
1WE5
 
 | | Crystal Structure of Alpha-Xylosidase from Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative family 31 glucosidase yicI | | Authors: | Ose, T, Kitamura, M, Okuyama, M, Mori, H, Kimura, A, Watanabe, N, Yao, M, Tanaka, I. | | Deposit date: | 2004-05-24 | | Release date: | 2005-02-15 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Alpha-Xylosidase from Escherichia coli
TO BE PUBLISHED
|
|
2UWM
 
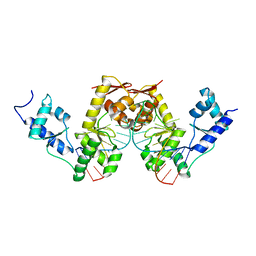 | | C-TERMINAL DOMAIN(WH2-WH4) OF ELONGATION FACTOR SELB IN COMPLEX WITH SECIS RNA | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*UP*GP*CP*CP*GP *GP*UP*CP*UP*GP*GP*CP*AP*AP*CP*GP*CP*C)-3', SELENOCYSTEINE-SPECIFIC ELONGATION FACTOR | | Authors: | Ose, T, Soler, N, Rasubala, L, Kuroki, K, Kohda, D, Fourmy, D, Yoshizawa, S, Maenaka, K. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis for Dynamic Interdomain Movement and RNA Recognition of the Selenocysteine-Specific Elongation Factor Selb.
Structure, 15, 2007
|
|
3A3Q
 
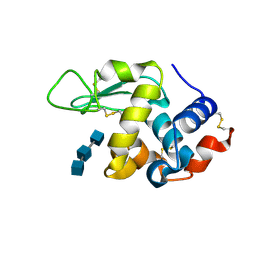 | | Structure of N59D HEN EGG-WHITE LYSOZYME in complex with (GlcNAc)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
3A3R
 
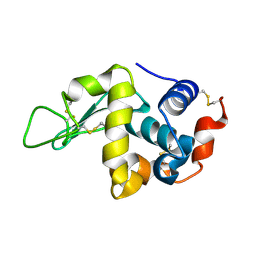 | | Structure of N59D HEN EGG-WHITE LYSOZYME | | Descriptor: | Lysozyme C | | Authors: | Ose, T, Kuroki, K, Matsushima, M, Maenaka, K, Kumagai, I. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of the hydrogen bonding network including Asp52 for catalysis, as revealed by Asn59 mutant hen egg-white lysozymes
J.Biochem., 146, 2009
|
|
1J0E
 
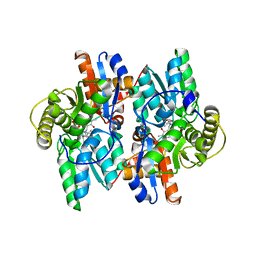 | | ACC deaminase mutant reacton intermediate | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
1IZC
 
 | | Crystal Structure Analysis of Macrophomate synthase | | Descriptor: | MAGNESIUM ION, PYRUVIC ACID, macrophomate synthase intermolecular Diels-Alderase | | Authors: | Ose, T, Watanabe, K, Mie, T, Honma, M, Watanabe, H, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into a natural Diels-Alder reaction from the structure of macrophomate synthase.
Nature, 422, 2003
|
|
1J0C
 
 | | ACC deaminase mutated to catalytic residue | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
5J7V
 
 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7O
 
 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein, unknown | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7U
 
 | | Faustovirus major capsid protein | | Descriptor: | major capsid protein, unknown | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of faustovirus, a large dsDNA virus.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7W71
 
 | |
5TIQ
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.537 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5TIP
 
 | | The Structure of the Major Capsid protein of PBCV-1 | | Descriptor: | 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose-(1-2)-beta-L-rhamnopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-D-mannopyranose-(1-3)-alpha-D-rhamnopyranose-(1-3)][alpha-D-galactopyranose-(1-2)]alpha-L-fucopyranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-glucopyranose, MERCURY (II) ION, Major capsid protein, ... | | Authors: | Klose, T, De Castro, C, Speciale, I, Molinaro, A, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the chlorovirus PBCV-1 major capsid glycoprotein determined by combining crystallographic and carbohydrate molecular modeling approaches.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6LA0
 
 | | Crystal structure of AoRut | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase family 5 | | Authors: | Koseki, T, Makabe, K. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aspergillus oryzae Rutinosidase: Biochemical and Structural Investigation.
Appl.Environ.Microbiol., 87, 2021
|
|
4Z25
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.339 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
4Z24
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
4Z26
 
 | | Mimivirus R135 (residues 51-702) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase R135 | | Authors: | Klose, T, Rossmann, M.G. | | Deposit date: | 2015-03-28 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.915 Å) | | Cite: | A Mimivirus Enzyme that Participates in Viral Entry.
Structure, 23, 2015
|
|
3WD4
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor and quinoline compound | | Descriptor: | (E)-N-(prop-2-en-1-yloxy)-1-(quinolin-4-yl)methanimine, Chitinase B, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD2
 
 | | Serratia marcescens Chitinase B complexed with azide inhibitor | | Descriptor: | Chitinase B, DITHIANE DIOL, GLYCEROL, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WD1
 
 | | Serratia marcescens Chitinase B complexed with syn-triazole inhibitor | | Descriptor: | Chitinase B, GLYCEROL, N,N-dibenzyl-N~5~-[N-(methylcarbamoyl)carbamimidoyl]-N~2~-{[5-({[(E)-(quinolin-4-ylmethylidene)amino]oxy}methyl)-1H-1,2,3-triazol-1-yl]acetyl}-L-ornithinamide, ... | | Authors: | Hirose, T, Maita, N, Gouda, H, Koseki, J, Yamamoto, T, Sugawara, A, Nakano, H, Hirono, S, Shiomi, K, Watanabe, T, Taniguchi, H, Sharpless, K.B, Omura, S, Sunazuka, T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Observation of the controlled assembly of preclick components in the in situ click chemistry generation of a chitinase inhibitor
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
