5WVU
 
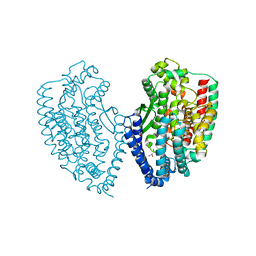 | | Crystal structure of carboxypeptidase from Thermus thermophilus | | Descriptor: | GLYCEROL, Thermostable carboxypeptidase 1, ZINC ION | | Authors: | Okai, M, Nagata, K, Tanokura, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the transition between the open and closed conformations of Thermus thermophilus carboxypeptidase.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
5TLC
 
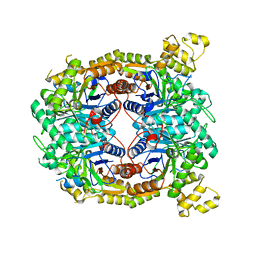 | |
2D37
 
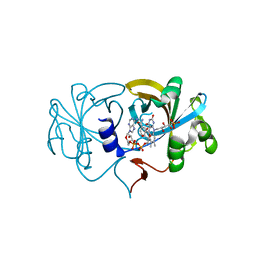 | | The Crystal Structure of Flavin Reductase HpaC complexed with NAD+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D38
 
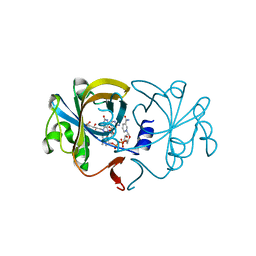 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
3A76
 
 | | The crystal structure of LinA | | Descriptor: | GLYCEROL, Gamma-hexachlorocyclohexane dehydrochlorinase, SPERMIDINE | | Authors: | Okai, M, Kubota, K, Fukuda, M, Nagata, Y, Nagata, K, Tanokura, M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of g-hexachlorocyclohexane dehydrochlorinase LinA from Sphingobium japonicum UT26
J.Mol.Biol., 2010
|
|
2YYS
 
 | | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8 | | Descriptor: | GLYCEROL, Proline iminopeptidase-related protein | | Authors: | Okai, M, Miyauchi, Y, Ebihara, A, Lee, W.C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-01 | | Release date: | 2008-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the proline iminopeptidase-related protein TTHA1809 from Thermus thermophilus HB8
Proteins, 70, 2008
|
|
4H7E
 
 | | Crystal structure of haloalkane dehalogenase LinB V112A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7F
 
 | | Crystal structure of haloalkane dehalogenase LinB V134I mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7D
 
 | | Crystal structure of haloalkane dehalogenase LinB T81A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H77
 
 | | Crystal structure of haloalkane dehalogenase LinB from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, F.L, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7J
 
 | | Crystal structure of haloalkane dehalogenase LinB H247A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7K
 
 | | Crystal structure of haloalkane dehalogenase LinB I253M mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7H
 
 | | Crystal structure of haloalkane dehalogenase LinB T135A mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
4H7I
 
 | | Crystal structure of haloalkane dehalogenase LinB L138I mutant from Sphingobium sp. MI1205 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Okai, M, Ohtsuka, J, Imai, L.F, Mase, T, Moriuchi, R, Tsuda, M, Nagata, K, Nagata, Y, Tanokura, M. | | Deposit date: | 2012-09-20 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Site-Directed Mutagenesis Analyses of Haloalkane Dehalogenase LinB from Sphingobium sp. Strain MI1205.
J.Bacteriol., 195, 2013
|
|
1ZOV
 
 | | Crystal Structure of Monomeric Sarcosine Oxidase from Bacillus sp. NS-129 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Nagata, K, Sasaki, H, Ohtsuka, J, Hua, M, Okai, M, Kubota, K, Kamo, M, Ito, K, Ichikawa, T, Koyama, Y, Tanokura, M. | | Deposit date: | 2005-05-14 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of monomeric sarcosine oxidase from Bacillus sp. NS-129 reveals multiple conformations at the active-site loop
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
5YPX
 
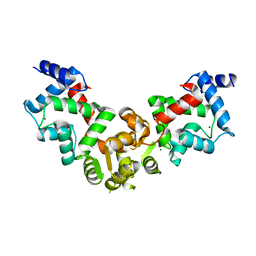 | | Crystal structure of calaxin with magnesium | | Descriptor: | Calaxin, MAGNESIUM ION | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-11-04 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
5X9A
 
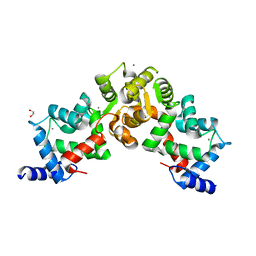 | | Crystal structure of calaxin with calcium | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calaxin | | Authors: | Shojima, T, Hou, F, Takahashi, Y, Okai, M, Mizuno, K, Inaba, K, Miyakawa, T, Tanokura, M. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a Ca2+-dependent regulator of flagellar motility reveals the open-closed structural transition
Sci Rep, 8, 2018
|
|
2Z1N
 
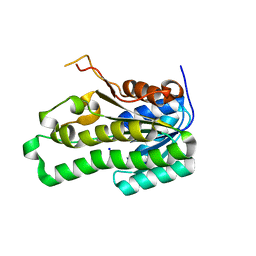 | | Crystal structure of APE0912 from Aeropyrum pernix K1 | | Descriptor: | SODIUM ION, dehydrogenase | | Authors: | Ichimura, T, Yamamura, A, Mimoto, F, Ohtsuka, J, Miyazono, K, Okai, M, Kamo, M, Lee, W.-C, Nagata, K, Tanokura, M. | | Deposit date: | 2007-05-10 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A unique catalytic triad revealed by the crystal structure of APE0912, a short-chain dehydrogenase/reductase family protein from Aeropyrum pernix K1
Proteins, 70, 2008
|
|
2ZUA
 
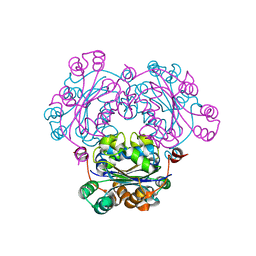 | | Crystal structure of nucleoside diphosphate kinase from Haloarcula quadrata | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Ichimura, T, Yamamura, A, Ohtsuka, J, Miyazono, K, Okai, M, Nagata, K, Tanokura, M. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular mechanism of distinct salt-dependent enzyme activity of two halophilic nucleoside diphosphate kinases
Biophys.J., 96, 2009
|
|
3AI3
 
 | | The crystal structure of L-Sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose | | Descriptor: | L-sorbose, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase, ... | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3AI2
 
 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-07 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3WGB
 
 | | Crystal structure of aeromonas jandaei L-allo-threonine aldolase | | Descriptor: | GLYCINE, L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3AI1
 
 | | The crystal structure of L-sorbose reductase from Gluconobacter frateurii complexed with NADPH and L-sorbose reveals the structure bases of its catalytic mechanism and high substrate selectivity | | Descriptor: | NADPH-sorbose reductase | | Authors: | Kubota, K, Nagata, K, Okai, M, Miyazono, K, Tanokura, M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The Crystal Structure of l-Sorbose Reductase from Gluconobacter frateurii Complexed with NADPH and l-Sorbose
J.Mol.Biol., 407, 2011
|
|
3WGC
 
 | | Aeromonas jandaei L-allo-threonine aldolase H128Y/S292R double mutant | | Descriptor: | L-allo-threonine aldolase, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE] | | Authors: | Qin, H.M, Imai, F.L, Miyakawa, T, Kataoka, M, Okai, M, Ohtsuka, J, Hou, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2013-08-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | L-allo-Threonine aldolase with an H128Y/S292R mutation from Aeromonas jandaei DK-39 reveals the structural basis of changes in substrate stereoselectivity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
