5GT6
 
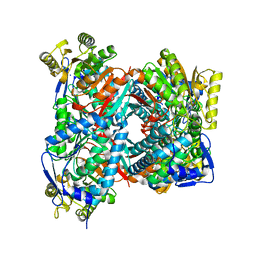 | | Apo structure of Aldehyde Dehydrogenase from Bacillus cereus | | Descriptor: | Betaine-aldehyde dehydrogenase, SODIUM ION | | Authors: | Ngo, H.P.T, Hong, S.H, Ho, T.H, Oh, D.K, Kang, L.W. | | Deposit date: | 2016-08-18 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | crystal structures of aldehyde dehydrogenase from Bacillus cereus having atypical bidirectional oxidizing and reducing activities for all-trans-retinal
To Be Published
|
|
5GTL
 
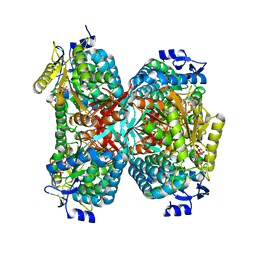 | | NADPH complex structure of Aldehyde Dehydrogenase from Bacillus cereus | | Descriptor: | Betaine-aldehyde dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION | | Authors: | Ngo, H.P.T, Hong, S.H, Ho, T.H, Oh, D.K, Kang, L.W. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of aldehyde dehydrogenase from Bacillus cereus having atypical bidirectional oxidizing and reducing activities for all-trans-retinal
To Be Published
|
|
5GTK
 
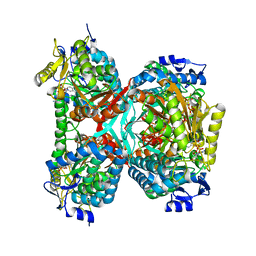 | | NAD+ complex structure of aldehyde dehydrogenase from bacillus cereus | | Descriptor: | Betaine-aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Ngo, H.P.T, Hong, S.H, Ho, T.H, Oh, D.K, Kang, L.W. | | Deposit date: | 2016-08-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of aldehyde dehydrogenase from Bacillus cereus having atypical bidirectional oxidizing and reducing activities for all-trans-retinal
To Be Published
|
|
3PH3
 
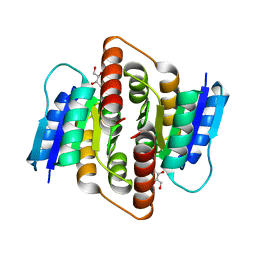 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-ribose | | Descriptor: | D-ribose, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.K, Yeom, S.J, Ahn, Y.J, Oh, D.K, Kang, L.W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
2HK0
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the absence of substrate | | Descriptor: | D-PSICOSE 3-EPIMERASE | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
4QF6
 
 | |
2HK1
 
 | | Crystal structure of D-psicose 3-epimerase (DPEase) in the presence of D-fructose | | Descriptor: | D-PSICOSE 3-EPIMERASE, D-fructose, MANGANESE (II) ION | | Authors: | Kim, K, Kim, H.J, Oh, D.K, Cha, S.S, Rhee, S. | | Deposit date: | 2006-07-03 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of d-Psicose 3-epimerase from Agrobacterium tumefaciens and its Complex with True Substrate d-Fructose: A Pivotal Role of Metal in Catalysis, an Active Site for the Non-phosphorylated Substrate, and its Conformational Changes
J.Mol.Biol., 361, 2006
|
|
4QET
 
 | |
