3ZTR
 
 | | Hexagonal form P6122 of the Aquifex aeolicus nucleoside diphosphate kinase (FIRST STAGE OF RADIATION DAMAGE) | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
3ZTP
 
 | | Orthorhombic crystal form P21212 of the Aquifex aeolicus nucleoside diphosphate kinase | | Descriptor: | GLYCEROL, NUCLEOSIDE DIPHOSPHATE KINASE, SULFATE ION | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An Intersubunit Disulfide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus.
Proteins, 80, 2012
|
|
3ZTQ
 
 | | Hexagonal crystal form P61 of the Aquifex aeolicus nucleoside diphosphate kinase | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Boissier, F, Georgescauld, F, Moynie, L, Dupuy, J.-W, Sarger, C, Podar, M, Lascu, I, Giraud, M.-F, Dautant, A. | | Deposit date: | 2011-07-12 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Inter-Subunit Disulphide Bridge Stabilizes the Tetrameric Nucleoside Diphosphate Kinase of Aquifex Aeolicus
Proteins, 80, 2012
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Z
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7P23
 
 | |
7P22
 
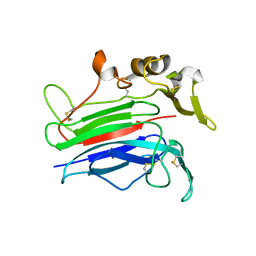 | |
8WS2
 
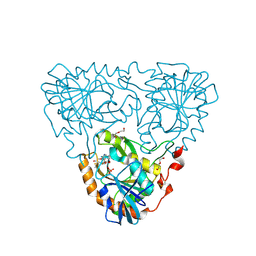 | | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Iizuka, Y, Kikuchi, M, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine
To Be Published
|
|
9JD2
 
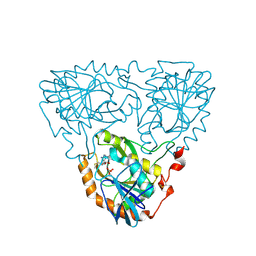 | |
9JHV
 
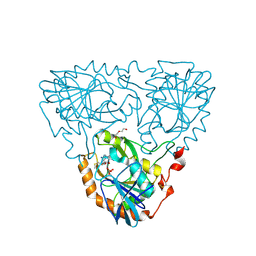 | | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 343K | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Iizuka, Y, Kikuchi, M, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2024-09-10 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 353K
To Be Published
|
|
5JQP
 
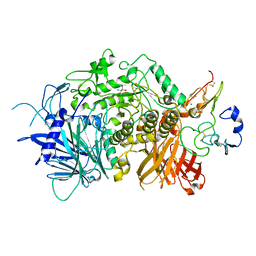 | | Crystal structure of ER glucosidase II heterodimeric complex consisting of catalytic subunit and the binding domain of regulatory subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha glucosidase-like protein, CALCIUM ION, ... | | Authors: | Satoh, T, Toshimori, T, Noda, M, Uchiyama, S, Kato, K. | | Deposit date: | 2016-05-05 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction mode between catalytic and regulatory subunits in glucosidase II involved in ER glycoprotein quality control.
Protein Sci., 25, 2016
|
|
4U4P
 
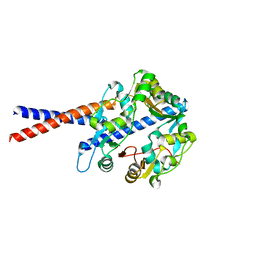 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
1FZZ
 
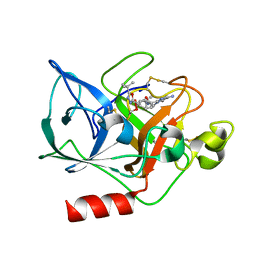 | | THE CRYSTAL STRUCTURE OF THE COMPLEX OF NON-PEPTIDIC INHIBITOR ONO-6818 AND PORCINE PANCREATIC ELASTASE. | | Descriptor: | 2-(5-AMINO-6-OXO-2-PHENYL-6H-PYRIMIDIN-1-YL)-N-[2-(5-TERT-BUTYL-1,3,4-OXADIAZOL-2-YL)-1-(METHYLETHYL)-2-HYDROXYETHYL]ACETAMIDE, ELASTASE 1 | | Authors: | Odagaki, Y, Ohmoto, K, Matsuoka, S, Hamanaka, N, Nakai, H, Toda, M, Katsuya, Y. | | Deposit date: | 2000-10-04 | | Release date: | 2001-10-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The crystal structure of the complex of non-peptidic inhibitor of human neutrophil elastase ONO-6818 and porcine pancreatic elastase.
Bioorg.Med.Chem., 9, 2001
|
|
3IS1
 
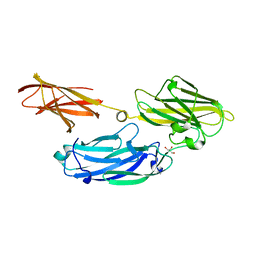 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in C2 form at 2.45 angstrom resolution | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Matsuoka, E, Shouji, Y, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IS0
 
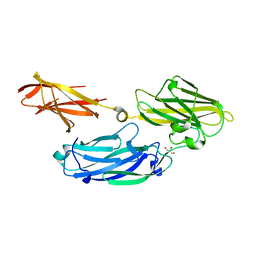 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in the presence of cholesterol | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Functional Region of UafA from Staphylococcus saprophyticus
To be Published
|
|
3IRP
 
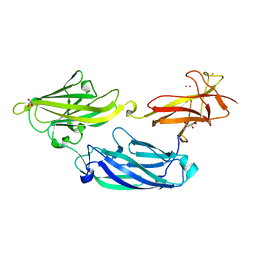 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus at 1.50 angstrom resolution | | Descriptor: | GLYCEROL, POTASSIUM ION, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
3IRZ
 
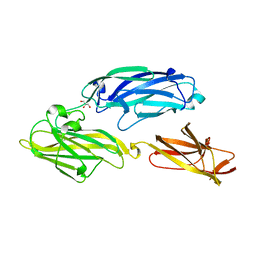 | | Crystal structure of functional region of UafA from Staphylococcus saprophyticus in P212121 form | | Descriptor: | GLYCEROL, Uro-adherence factor A | | Authors: | Tanaka, Y, Shouji, Y, Matsuoka, E, Kuroda, M, Tanaka, I, Yao, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the functional region of Uro-adherence factor A from Staphylococcus saprophyticus reveals participation of the B domain in ligand binding
Protein Sci., 20, 2011
|
|
5Y0D
 
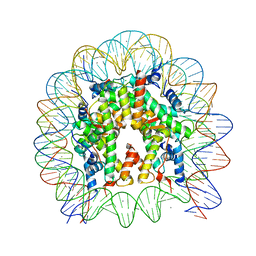 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | Deposit date: | 2017-07-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
7VMR
 
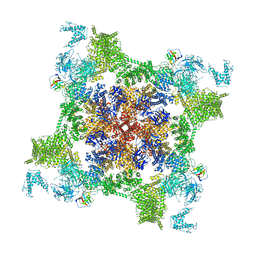 | | Structure of recombinant RyR2 mutant K4593A (EGTA dataset) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMM
 
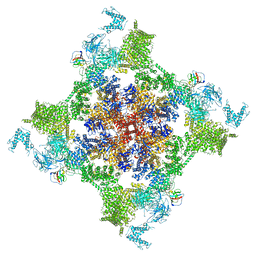 | | Structure of recombinant RyR2 (EGTA dataset, class 1, closed state) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ZINC ION | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations.
Nat Commun, 13, 2022
|
|
7VMS
 
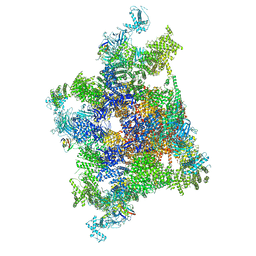 | | Structure of recombinant RyR2 mutant K4593A (Ca2+ dataset) | | Descriptor: | CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, Ryanodine receptor 2, ... | | Authors: | Kobayashi, T, Tsutsumi, A, Kurebayashi, N, Kodama, M, Kikkawa, M, Murayama, T, Ogawa, H. | | Deposit date: | 2021-10-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations
Nat Commun, 13, 2022
|
|
