7Z7U
 
 | | REP-related Chom18 variant with double CG base pair | | Descriptor: | Chom18-CG DNA, STRONTIUM ION | | Authors: | Svoboda, J, Kolenko, P, Berdar, D, Schneider, B. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7L
 
 | | REP-related Chom18 variant with double AC mismatch | | Descriptor: | Chom18-AC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7Y
 
 | | REP-related Chom18 variant with double GT mismatch | | Descriptor: | Chom18-GT DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7Z
 
 | | REP-related Chom18 variant with double TA base pair | | Descriptor: | Chom18-TA DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7M
 
 | | REP-related Chom18 variant with double CC mismatch | | Descriptor: | Chom18-CC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z7K
 
 | | REP-related Chom18 variant with double AT base pairing | | Descriptor: | Chom18-AT DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7Z81
 
 | | REP-related Chom18 variant with double TC mismatch | | Descriptor: | Chom18-TC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
1GSH
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 7.5 | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Kato, H, Yamaguchi, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2AE2
 
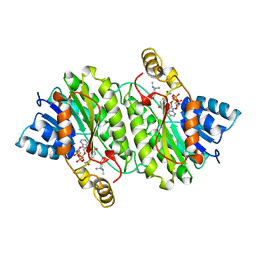 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADP+ AND PSEUDOTROPINE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (TROPINONE REDUCTASE-II), PSEUDOTROPINE | | Authors: | Yamashita, A, Kato, H, Wakatsuki, S, Tomizaki, T, Nakatsu, T, Nakajima, K, Hashimoto, T, Yamada, Y, Oda, J. | | Deposit date: | 1999-01-26 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of tropinone reductase-II complexed with NADP+ and pseudotropine at 1.9 A resolution: implication for stereospecific substrate binding and catalysis.
Biochemistry, 38, 1999
|
|
2GLT
 
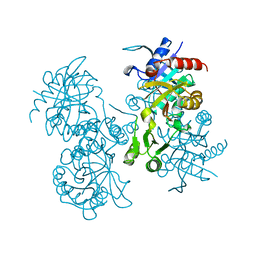 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2AE1
 
 | | TROPINONE REDUCTASE-II | | Descriptor: | TROPINONE REDUCTASE-II | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1GSA
 
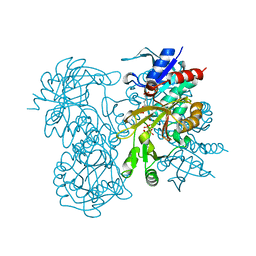 | | STRUCTURE OF GLUTATHIONE SYNTHETASE COMPLEXED WITH ADP AND GLUTATHIONE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, GLUTATHIONE SYNTHETASE, ... | | Authors: | Hara, T, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-06-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution.
Biochemistry, 35, 1996
|
|
1IPF
 
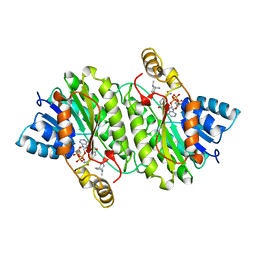 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH AND TROPINONE | | Descriptor: | 8-METHYL-8-AZABICYCLO[3,2,1]OCTAN-3-ONE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1IPE
 
 | | TROPINONE REDUCTASE-II COMPLEXED WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-II | | Authors: | Yamashita, A, Endo, M, Higashi, T, Nakatsu, T, Yamada, Y, Oda, J, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-05-09 | | Release date: | 2003-06-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing Enzyme Structure Prior to Reaction Initiation: Tropinone Reductase-II-Substrate Complexes
BIOCHEMISTRY, 42, 2003
|
|
1V4G
 
 | | Crystal Structure of gamma-Glutamylcysteine Synthetase from Escherichia coli B | | Descriptor: | Glutamate--cysteine ligase | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2003-11-13 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1VA6
 
 | | Crystal structure of Gamma-glutamylcysteine synthetase from Escherichia Coli B complexed with Transition-state analogue | | Descriptor: | (2S)-2-AMINO-4-[[(2R)-2-CARBOXYBUTYL](PHOSPHONO)SULFONIMIDOYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
11AS
 
 | |
12AS
 
 | | ASPARAGINE SYNTHETASE MUTANT C51A, C315A COMPLEXED WITH L-ASPARAGINE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE, ASPARAGINE SYNTHETASE | | Authors: | Nakatsu, T, Kato, H, Oda, J. | | Deposit date: | 1997-12-02 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of asparagine synthetase reveals a close evolutionary relationship to class II aminoacyl-tRNA synthetase.
Nat.Struct.Biol., 5, 1998
|
|
1AUN
 
 | | PATHOGENESIS-RELATED PROTEIN 5D FROM NICOTIANA TABACUM | | Descriptor: | PR-5D | | Authors: | Koiwa, H, Kato, H, Nakatsu, T, Oda, J, Yamada, Y, Sato, F. | | Deposit date: | 1997-08-29 | | Release date: | 1998-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tobacco PR-5d protein at 1.8 A resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins.
J.Mol.Biol., 286, 1999
|
|
1J2G
 
 | |
1AE1
 
 | | TROPINONE REDUCTASE-I COMPLEX WITH NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-I | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-23 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2D33
 
 | | Crystal Structure of gamma-Glutamylcysteine Synthetase Complexed with Aluminum Fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CYSTEINE, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of efficient coupling between peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
2D32
 
 | | Crystal Structure of Michaelis Complex of gamma-Glutamylcysteine Synthetase | | Descriptor: | CYSTEINE, GLUTAMIC ACID, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nakayama, M, Nii, H, Kurokawa, Y, Katano, H, Oda, J. | | Deposit date: | 2005-09-25 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of efficient coupling peptide ligation and ATP hydrolysis by gamma-gluatamylcysteine synthetase
To be Published
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QKA
 
 | | Crystal structure of SARS-CoV-2 Main Protease in complex with covalently bound GC376 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
