2E7U
 
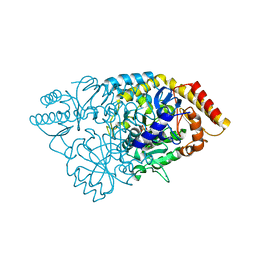 | | Crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from Thermus thermophilus HB8 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, GLYCEROL, Glutamate-1-semialdehyde 2,1-aminomutase, ... | | Authors: | Mizutani, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-14 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from Thermus thermophilus HB8
To be Published
|
|
2EHT
 
 | |
2EPI
 
 | |
2DZ9
 
 | |
2EKB
 
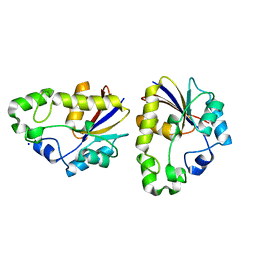 | | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M) | | Descriptor: | Alpha-ribazole-5'-phosphate phosphatase, SODIUM ION | | Authors: | Asada, Y, Taketa, M, Tanaka, Y, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-22 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural study of Project ID TTHB049 from Thermus thermophilus HB8 (L19M)
To be Published
|
|
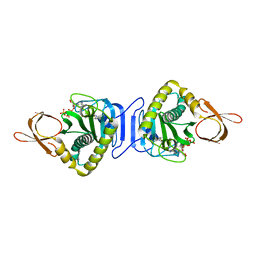
2DXT
 
 | |
2EO4
 
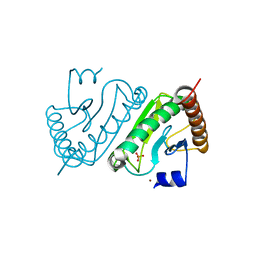 | |
2EVB
 
 | |
3AH9
 
 | | Crystal structure of (Pro-Pro-Gly)9 at 1.1 A resolution | | Descriptor: | ACETIC ACID, AZIDE ION, collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Hosaka, N, Nishino, N. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of (Pro-Pro-Gly)9
To be Published
|
|
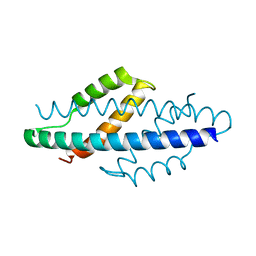
2D8E
 
 | |
2D5D
 
 | |
2D3K
 
 | |
2CUY
 
 | | Crystal structure of malonyl CoA-acyl carrier protein transacylase from Thermus thermophilus HB8 | | Descriptor: | Malonyl CoA-[acyl carrier protein] transacylase | | Authors: | Misaki, S, Suzuki, K, Kunishima, N, Sugawara, M, Kuroishi, C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-31 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of malonyl CoA-acyl carrier protein transacylase from Thermus thermophilus HB8
TO BE PUBLISHED
|
|
2DPL
 
 | |
2DQW
 
 | |
3W59
 
 | |
2D8D
 
 | |
3W58
 
 | |
2DJ6
 
 | |
2DJZ
 
 | |
2HIA
 
 | |
2DZC
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii, Mutation R48A | | Descriptor: | biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2HNI
 
 | |
2E64
 
 | |
2EGG
 
 | |
