1SV6
 
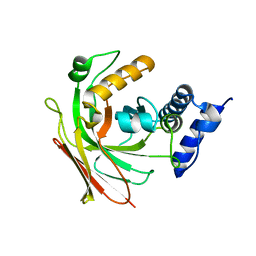 | | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli | | Descriptor: | 2-keto-4-pentenoate hydratase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sharp, A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-03-28 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of 2-hydroxypentadienoic acid hydratase from Escherichia Coli
To be Published
|
|
1YW1
 
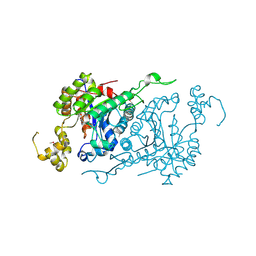 | |
2RDY
 
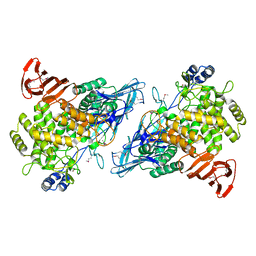 | |
2RJO
 
 | |
1X94
 
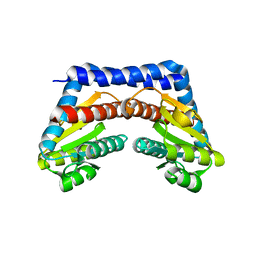 | |
1T62
 
 | |
2A1V
 
 | |
1ZWL
 
 | |
1WV2
 
 | |
2RAG
 
 | | Crystal structure of aminohydrolase from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, Dipeptidase, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of aminohydrolase from Caulobacter crescentus.
To be Published
|
|
1UB4
 
 | |
2RJN
 
 | | Crystal structure of an uncharacterized protein Q2BKU2 from Neptuniibacter caesariensis | | Descriptor: | Response regulator receiver:Metal-dependent phosphohydrolase, HD subdomain | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-15 | | Release date: | 2007-10-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an uncharacterized protein Q2BKU2 from Neptuniibacter caesariensis.
To be Published
|
|
1TK9
 
 | | Crystal Structure of Phosphoheptose isomerase 1 | | Descriptor: | Phosphoheptose isomerase 1 | | Authors: | Rajashankar, K.R, Solorzano, V, Kniewel, R, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-08 | | Release date: | 2004-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of two putative phosphoheptose isomerases.
Proteins, 63, 2006
|
|
1U6L
 
 | |
1RZN
 
 | |
1RGX
 
 | | Crystal Structure of resisitin | | Descriptor: | Resistin | | Authors: | Patel, S.D, Rajala, M.W, Scherer, P.E, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | Disulfide-dependent multimeric assembly of resistin family hormones
Science, 304, 2004
|
|
1TYH
 
 | |
1YRZ
 
 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
1YMY
 
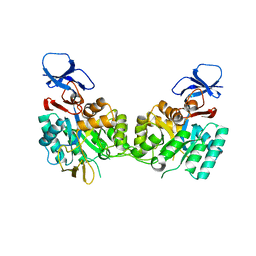 | | Crystal Structure of the N-Acetylglucosamine-6-phosphate deacetylase from Escherichia coli K12 | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the N-acetylglucosamine-6-phosphate deacetylase from Escherichia Coli
To be Published
|
|
1S7J
 
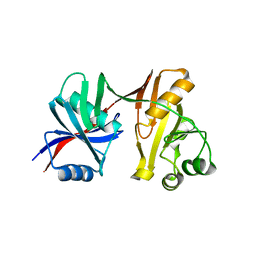 | |
1YWK
 
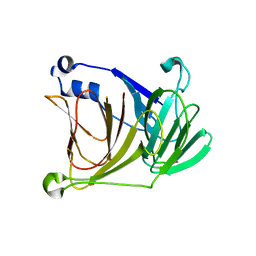 | |
1TVL
 
 | |
1Z4E
 
 | |
2OQY
 
 | | The crystal structure of muconate cycloisomerase from Oceanobacillus iheyensis | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Toro, R, Fedorov, E.V, Bonanno, J, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
1YBF
 
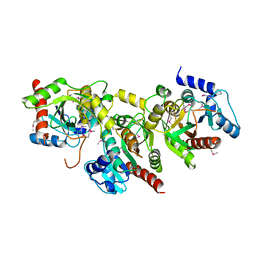 | |
