3S0C
 
 | | Transaldolase wt of Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1W
 
 | | Transaldolase variant Lys86Ala from Thermoplasma acidophilum in complex with glycerol and citrate | | Descriptor: | CITRATE ANION, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3S1V
 
 | | Transaldolase from Thermoplasma acidophilum in complex with D-fructose 6-phosphate Schiff-base intermediate | | Descriptor: | FRUCTOSE -6-PHOSPHATE, GLYCEROL, Probable transaldolase | | Authors: | Lehwess-Litzmann, A, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Twisted Schiff base intermediates and substrate locale revise transaldolase mechanism.
Nat.Chem.Biol., 7, 2011
|
|
3V0X
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bovine trypsin variants X(tripleGlu217Phe227) in complexes with small molecule inhibitor
To be Published
|
|
7NZJ
 
 | | Structure of bsTrmB apo | | Descriptor: | GLYCEROL, SODIUM ION, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NZI
 
 | | TrmB complex with SAH | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
7NYB
 
 | | TrmB complex with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(7)-)-methyltransferase | | Authors: | Blersch, K.F, Ficner, R, Neumann, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural model of the M7G46 Methyltransferase TrmB in complex with tRNA.
Rna Biol., 18, 2021
|
|
6HVN
 
 | | CdaA-APO Y187A Mutant | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.234 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
6HVL
 
 | | CdaA complex with c-di-AMP and AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
6H45
 
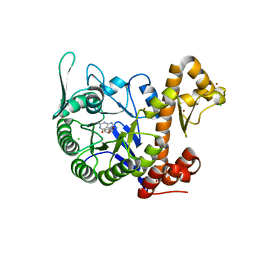 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
6H42
 
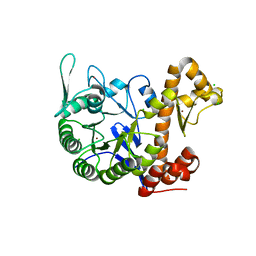 | | crystal structure of the human TGT catalytic subunit QTRT1 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
4IA6
 
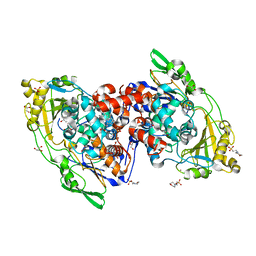 | | Hydratase from lactobacillus acidophilus in a ligand bound form (LA LAH) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6HVM
 
 | | Structural characterization of CdaA-APO | | Descriptor: | CHLORIDE ION, Diadenylate cyclase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Heidemann, J.L, Neumann, P, Ficner, R. | | Deposit date: | 2018-10-11 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the c-di-AMP-synthesizing enzyme CdaA.
J.Biol.Chem., 294, 2019
|
|
4HZK
 
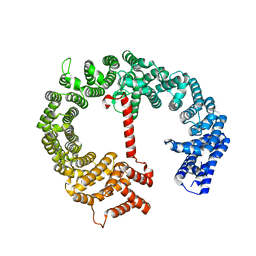 | |
4IA5
 
 | | Hydratase from Lactobacillus acidophilus - SeMet derivative (apo LAH) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Khoshnevis, S, Neumann, P, Ficner, R. | | Deposit date: | 2012-12-06 | | Release date: | 2013-03-27 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis of a fatty acid double-bond hydratase from Lactobacillus acidophilus
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JQ9
 
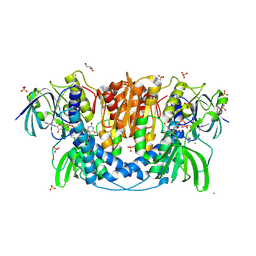 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
6FA9
 
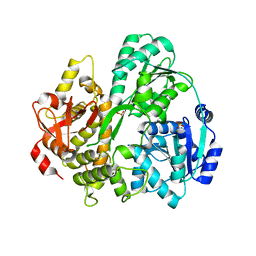 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 | | Descriptor: | Putative mRNA splicing factor, SULFATE ION | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FDF
 
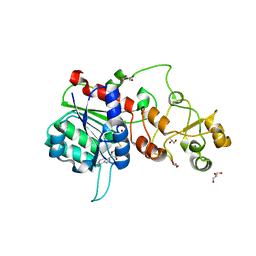 | |
6F2C
 
 | | Methylglyoxal synthase MgsA from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, Methylglyoxal synthase | | Authors: | Dickmanns, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-11-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the regulatory interaction of the methylglyoxal synthase MgsA with the carbon flux regulator Crh inBacillus subtilis.
J. Biol. Chem., 293, 2018
|
|
6FAC
 
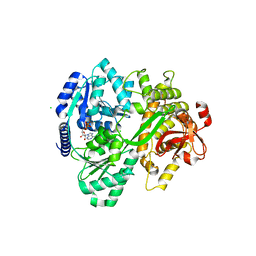 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FA5
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hamann, F, Schmitt, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5MEE
 
 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304V | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arachidonate 15-lipoxygenase, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5MEG
 
 | | Manganese-substituted Cyanothece lipoxygenase 2 (Mn-CspLOX2) | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, ETHANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5O5O
 
 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
